HBV transcription
Abstract: The X mRNA seems to be transcribed early and transiently, whereas structural proteins mRNAs are on the contrary expressed later and constantly. The S promoter would thus be more active that the PreS1, maybe explaining the huge ammount of SHB and MHB proteins synthesized during infection.
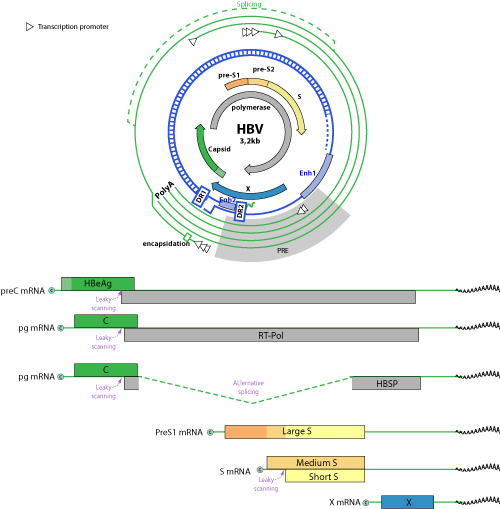
Early and late transcription
HBV transcription is under the control of four promoters (the core, pre-S1, pre-S2/S, and X promoters) and two enhancer regions (ENH1 and ENH2). Doitsh and colleagues showed that enhancer I was responsible for the activation of the early transcripts (X mRNA) while enhancer II was involved in the expression of the late transcripts (pg, pc, and S mRNAs) (Doitsh et al. 2004) Rating=2. The early transcripts HBx mRNA accumulates early after transfection (about 16 hours post transfection) and disappear after 4 days (Wu et al. 1991, Doitsh et al. 2003) Rating=2. All the other HBV RNAs are referred as late transcripts and accumulate about 20 hours post transfection.
HBV tropism is probably restricted at the transcriptional level by the need for hepatic nuclear receptors.
Transcription from the X promoter
Two RNAs are synthesized from the X promoter: a short one termed sx (0.7kb) and a long one termed lx (3.9kb), the latter being produced when the polymerase does not stop at the polyA signal and thus transcribes an additional genome's round (figure in Doitsh et al. 2004). This long transcript does not seem to be efficiently exported from the nucleus (Doitsh et al. 2003) Rating=2.
HBx promoter has been shown to be under control of the nuclear respiratory factor 1 (NRF1), an activator of mitochondrial genes (Tokusumi et al. 2004) Rating=1.
Transcription from the pc/pgRNA promoters
The pre-genomic (3.5kb) and precore (3.4kb) transcripts are produced from distinct juxtaposed promoter sequences (Yu et al. 1996) Rating=1. For example, the hepatocyte nuclear factor 4 (HNF-4) modestly affects transcription from the precore promoter while it strongly activates transcription from the pgRNA promoter (Yu et al. 2003) Rating=1.
Transcription from the preS1 promoter
The preS1 promoter is located upstream the preS1 mRNA which encodes for the large surface envelop protein of HBV. The transcription factor HNF1alpha regulates the expression of the 2.4 kb RNA. The HNF-1 site in the HBV genome is important for hepatocyte-specific activity of the preSl promoter but not of the S promoter. By itself, HNF-1 is not able to activate transcription of the HBV preS1 promoter, but requires also the presence of the Oct-1 cofactor to be efficient (Zhou et al. 1991) Rating=1.
Transcription from the S promoter
Transcription from the S promoter (producing the RNA encoding for middle and small envelope proteins) would be responsive to ubiquitous transcription factors in vitro (Raney et al. 1992) Rating=2. It is also clearly responsive to RXR-alpha (Tang et al. 2001) Rating=2 and HNF-4 (Doitsh et al. 2004) Rating=2. The ratio observed for the transcription of S versus preS RNAs is about 1/5 (Bock et al. 1999) Rating=1.
In vivo however, tissue distribution of the 2.1 kb HBV transcript is primarily restricted to the liver and kidney of HBV transgenic mice. Its expression seems thus to depend on liver-specific factors, e.g. HNF-3 (Raney et al. 1997) Rating=2.
Transcription termination
The pgRNA size is greater than the genome size itself (3.5 vs 3.2 kb). The transcribing cellular polymerase ignores the polyA/stop signal which is present 300bp away from the start, but the same site is efficiently recognized when the RNA is 3.5kb. Possibly the host RNA-polymerase II is in an activated state during transcription of the first hundreds bp. This would prevent pausing and termination factors function (Guo et al. 2011) Rating=2. This model would explain why HBx mRNA can be short or long (sxRNA and lxRNA): the polymerase would still be not totally 100% competent to stop after 700 bp.