Eukaryotic host translation shutoff by virus (kw:KW-1193)
The majority of cellular mRNAs initiate translation through the recruitment of a multisubunit translation initiation complex termed eIF4F, which consists of the cap-binding protein eIF4E, the RNA helicase eIF4A, and the adaptor protein eIF4G. eIF4G binds poly(A)-binding protein (PABP) to mediate 5' -> 3' communication, probably to promote efficient translation of intact correctly processed mRNAs.
eIF4E binding protein 1 (eIF4E-BP1) functions as a translational repressor that limits eIF4E availability and therefore eIF4F complex formation.
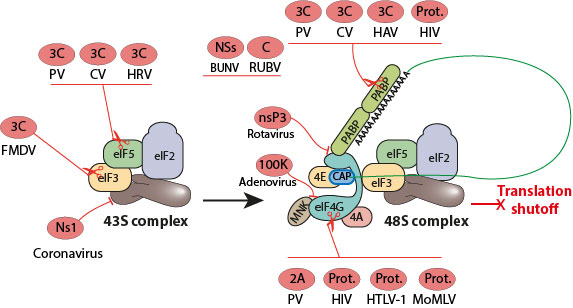
Viruses have evolved ways of interacting with the host translational machinery to shutoff host gene expression. This global inhibition of cellular protein synthesis serves to ensure maximal viral gene expression and to evade host immune response

 .
.
All the viruses inducing the shutoff of translation are able to continue to translate at least part of their mRNAs using non-canonical translation: IRES, Ribosome shunting, or VPG initiation. For adenoviridae, polyomaviridae and togaviridae the cellular translation shutoff takes place at the late phase of infection and ensures an optimal production of viral structural proteins.
Under stress conditions such as viral infection some cellular translation can continue when the cap-dependent translation initiation is inhibited. Expression of specific cellular proteins seems to occur through cap-independent mechanisms

 .
.
Viruses inducing host translation shutoff:
| Family | Virus | Viral protein | Shutoff strategy | Viral translation | ref. |
| Adenoviridae | Adenovirus | Shutoff protein 100K | Inhibits eIF4G-Mnk1 and prevents EIF4E phosphorylation | Ribosome shunting on late mRNAs |


|
| Bunyaviridae | Bunyamwera virus | Nucleoprotein | Nuclear retention of PABP | Viral mRNAs non polyadenylated | 
|
| Caliciviridae | Norovirus Feline calicivirus | Protease 3C Protease |
PABP cleavage | VPg
 |

|
| Feline calicivirus | Protease | Cleavage of eIF4G1 and eIF4G2 | VPg  |
 |
|
| Dicistroviridae | Cripavirus | ? | Dissociation of eIF4G and eIF4E | IRES |

|
| Orthomyxoviridae | Influenza virus | ? | Dephosphorylation of of eIF-4E | eIF-4E independent translation of viral mRNAs |


|
| Picornaviridae | Poliovirus | Protease 2A | Cleavage of eIF4GI and eIF4GII | IRES |


|
| Protease 3C | Cleavage of eIF5B | IRES |

|
||
| Protease 3C | PABP cleavage | IRES |

|
||
| Rhinovirus | Protease 2A | Cleavage of eIF4GI and eIF4GII | IRES |


|
|
| Coxsackie virus | Protease 2A | Cleavage of eIF4GI and eIF4GII | IRES |


|
|
| Coxsackie virus | Protease 2A,3C | PABP cleavage | IRES |


|
|
| Protease 3C | Cleavage of eIF5B | IRES |

|
||
| Encephalomyocarditis virus | Protein 2A | ? | IRES |

|
|
| ? | Dephosphorylation of 4E-BP1 | IRES |


|
||
| Foot-and-mouth disease virus | Leader protease | Cleavage of eIF4GI and eIF4GII | IRES |

 |
|
| Protease 3C | Cleavage of PABP, eIF3a, eIF3b, and eIF4A | IRES |

|
||
| Polyomaviridae | SV40 | Small T antigen | Dephosphorylation of 4E-BP1 | IRES |


|
| Reoviridae | Rotavirus | NSP3 | Interacts with EIF4G and evicts PABP from initiation complexes. Nuclear relocalization of PABP | NSP3 replaces PABP and bind the non polyAdenylated viral mRNAs |


|
| Rhabdoviridae | Vesicular stomatitis virus | ? | Dephosphorylation of 4E-BP1 | classical |

|
| Retroviridae | HIV-1 | Protease | Cleavage of eIF4GI and PABP | IRES |



|
| HTLV-1 | Protease | Cleavage of eIF4GI | IRES |

|
|
| Moloney murine leukemia virus | Protease | Cleavage of eIF4GI and eIF4GII | IRES |

|
|
| Togaviridae | Rubella virus | Capsid protein | PABP sequestration | classical |

|
| Sindbis virus | dsRNA leading to PKR-mediated inhibition of eIF2alpha | DLP |


| ||
| Coronaviridae | SARS-CoV | Nsp1 | Binding to host 40S subunit | Viral mRNA protected by 5'-end leader sequence |


|
| Bat-CoV | Nsp1 | - | - |

|
|
| TGEV | Nsp1 | - | - |

|
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)0 entry grouped by protein
