Viral genome circularization (kw:KW-1253)
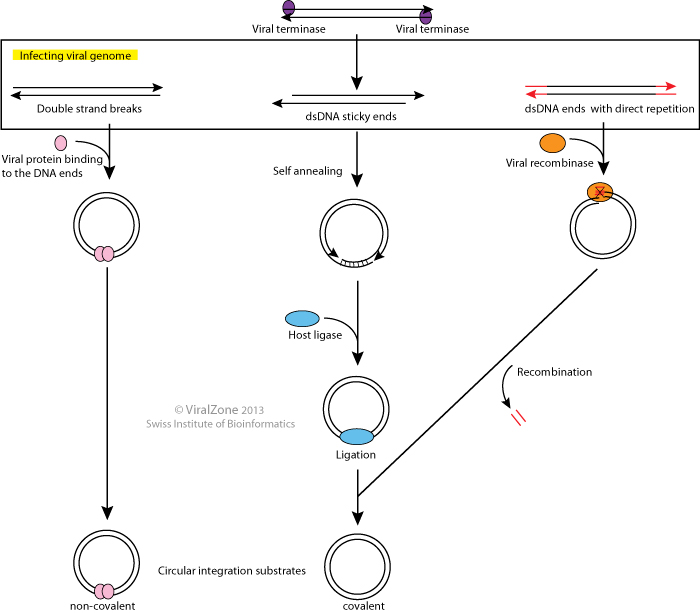
Circularization of infecting DNA within the host cell is a rather common amongst bacterial viruses to protect the viral genome ends from nucleases, to convert the linear genome to an integrative precursor or to give rise to the replicative form of the genome.
Circularization, also known as cyclization, can be mediated by covalent closure of the DNA "sticky" ends, recombinaison between redundant terminal sequences or via the binding of a protein at the viral DNA extremities.
| Virus | Family | Circularization mechanism | Viral protein | Ref. |
| Phage Mu | Myoviridae | Circularization protein (non-covalent) | Protein N |  |
| Phage lambda | Siphoviridae | Cohesive (sticky) ends | - | |
| Phage P22 | Podoviridae | Recombinaison | - |  Recombinational circularization of Salmonella phage P22 DNA S Weaver, M Levine Virology January 1977; 76: 29-38 |
| Phage P1 | Myoviridae | Recombinaison | Cre |  Genome of bacteriophage P1 Ma?gorzata B ?obocka, Debra J Rose, Guy Plunkett 3rd, Marek Rusin, Arkadiusz Samojedny, Hansj?rg Lehnherr, Michael B Yarmolinsky, Frederick R Blattner J. Bacteriol. November 2004; 186: 7032-7068 |
| Phage N15 | Siphoviridae | Cohesive (sticky) ends | - |  Conversion of linear DNA with hairpin telomeres into a circular molecule in the course of phage N15 lytic replication Mardanov AV, Ravin NV J Mol Biol. August 2009; 391: 261-8 |
Bacteriophage Mu DNA circularizes following infection of Escherichia coli
A H Puspurs, N J Trun, J N Reeve
EMBO J. 1983; 2: 345?352
A H Puspurs, N J Trun, J N Reeve
EMBO J. 1983; 2: 345?352
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)6 entries grouped by strain
1 entry
Enterobacteria phage GA (Bacteriophage GA) reference strain
1 entry
Escherichia phage MS2 (Bacteriophage MS2) reference strain
1 entry
Escherichia phage Mu (Bacteriophage Mu) reference strain
1 entry
Mycobacterium phage Corndog (Mycobacteriophage Corndog) reference strain
1 entry
Mycobacterium phage Omega (Mycobacteriophage Omega) reference strain
1 entry
