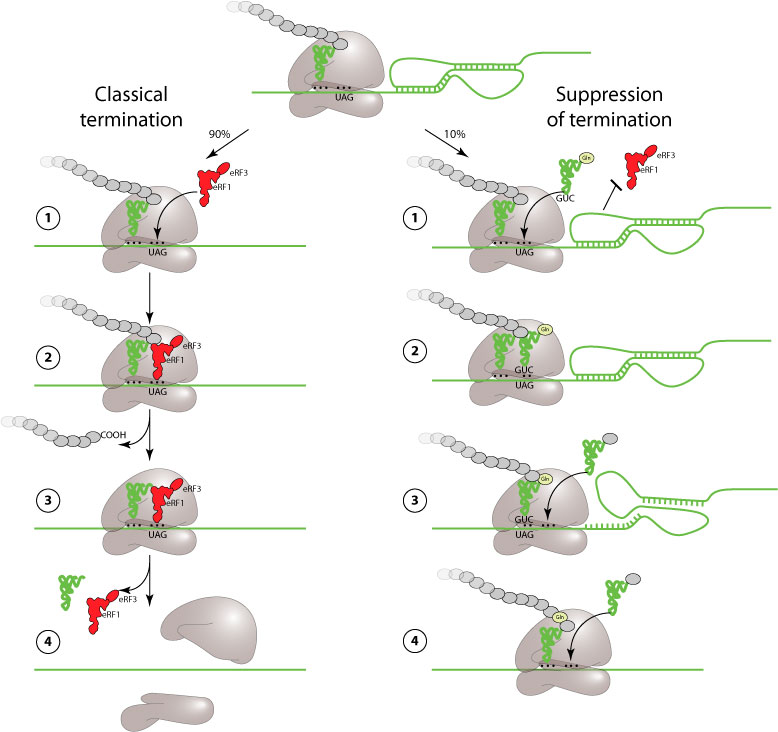
Suppression of termination (also referred to as stop codon readthrough) is an alternate mechanism of translation to produce a protein with an extended C-terminus. It is used as a regulatory strategy by a large number of plant, animal and prokaryot viruses (see table below).
UAG amber and UGA opal codons are normally recognized by a release factor (eRF) instead of a tRNA, thereby stopping translation. Readthrough of these stop codons occurs when a tRNA partially recognizes UGA or UAG and allows translation to proceed further. This readthrough is induced either by a pseudoknot (MMTV) or by few nucleotides sequence after the stop. The readthrough signals of prokaryote, plant and mammal viruses are different and have different mechanisms which are still poorly understood. The efficiency of suppression of termination is globally poor: 5 to 30%.
The picture below describes MMTV suppression of termination.