Virosaurus

Exploring viral genetic diversity for vertebrate metagenomics
Virosaurus (from virus thesaurus) is a curated virus genome database, aimed at facilitating clinical metagenomics analysis. The data comprises clustered and annotated sequences of Vertebrate viruses , Others viruses (Insect, Fungus, Eukaryotic microorgansism) or Plant viruses in FASTA format. Virosaurus also provides complete virus sequence dataset for all those viruses, which comprises complete genomes for nonsegmented viruses, and complete segments for segmented viruses.
beta version Virosaurus data browser
Download last version
| Name | Release | Complete virus sequences | Virosaurus90 | Virosaurus98 | User_Manual |
| Virosaurus databases 2020_4.2 | April 2020 | Complete 2020_4.1 | Vertebrate:(23615 FASTA) Virosaurus90v 2020_4.2 Others (insect, fungus,...):(9140 FASTA) Virosaurus90n 2020_4.1 Plants:(2786 FASTA) Virosaurus90p 2020_4.1 | Vertebrate:(73160 FASTA) Virosaurus98v 2020_4.2 Others (insect, fungus,...):(11971 FASTA) Virosaurus98n 2020_4.1 Plants:(6038 FASTA) Virosaurus98p 2020_4.1 | User Manual |
Description
Complete sequences: This dataset contains full-length genomes (monopartite virus) or segments (segmented virus) for all vertebrate virus families.
Virosaurus: Virus reference sequence databases for clinical metagenomics. All complete sequences were clustered at 90% to remove redundancy in Virosaurus Vertebrate 90 (23,615 FASTAs); or clustered at 98% in Virosaurus vertebrate 98 (73,160 FASTAs). Many clusters can belong to the same virus species. For example, there are 100 Lassa virus clusters in Virosaurus90, 638 in Virosaurus98.
The FASTA header have been annotated with metadata to facilitate metagenomic analysis. For instance, viral nucleic acid is annotated as RNA, DNA or RNA/DNA, thereby improving interpretation from sequencing either molecule.
Figure 1: Examples of virus genome annotation. The Usual name and clinical typing should be the default output for clinical studies.
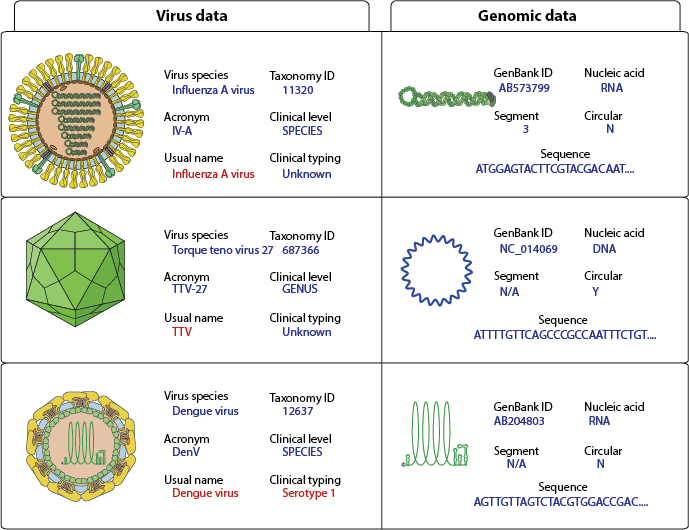 In the Virosaurus release 2019_10, herpesviridae and poxviridae sequences are split in genes rather than full genomes. This allows using incomplete genome sequences, and helps to mitigate the low number of complete genomes versus high variability for those families.
In the Virosaurus release 2019_10, herpesviridae and poxviridae sequences are split in genes rather than full genomes. This allows using incomplete genome sequences, and helps to mitigate the low number of complete genomes versus high variability for those families.
We suggest using Virosaurus90 if you need to optimize data analysis. If database size is not a problem, Virosaurus98 could be used for better resolution.
Archive
| Name | Release | Complete sequences | Virosaurus90 | Virosaurus98 | User_Manual | ||||
| Vertebrate | Others(Insects, fungus,...) | Plant | Vertebrate | Others(Insects, fungus,...) | Plant | ||||
| Virosaurus databases 2019_10.2 | Update January 2020 | Complete 2019_10.2 | V90v_2019_10.2 | V90n_2019_10 | V98v_2019_10.2 | V98n_2019_10 | User Manual | ||
| Version note: Removed LR584255 contaminant sequences | |||||||||
| Patch update 13 Janv 2020: Wuhan coronavirus fasta | |||||||||
|
|
|||||||||
| Virosaurus databases 2019_10 | October 2019 | Complete 2019_10 | V90v_2019_10 | V90n_2019_10 | V98v_2019_10 | V98n_2019_10 | User Manual | ||
|
|
|||||||||
| Virosaurus databases 2018_11 | November 2018 | Complete 2018_11 | V90v_2018_11 | V98v_2018_11 | User manual | ||||
Licence
 Attribution-NonCommercial-NoDerivatives 4.0 International
Attribution-NonCommercial-NoDerivatives 4.0 International
Authors
Anne Gleizes, Florian Laubscher, Nicolas Guex, Christian Iseli, Thomas Junier, Samuel Cordey, Jacques Fellay, Ioannis Xenarios, Laurent Kaiser and Philippe Le Mercier.
Credits
Virosaurus has been developed by a collaboration between SIB Swiss Institute of Bioinformatics (Vital-IT and Swiss-Prot groups), and Virology Laboratory of Geneva University Hospitals (HUG). The development of Virosaurus is supported by the Swiss National Science Foundation (grant 310030_189179).



