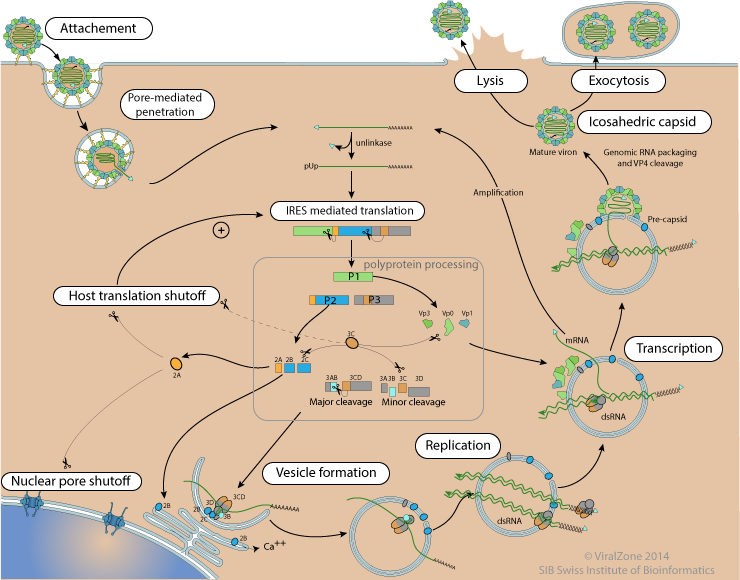
Poliovirus replication cycle
|
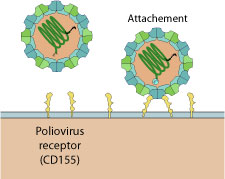
Poliovirus virion binds to host Pvr receptor (CD155) via depressions on capsid surface. Virion binding to receptors triggers endocytosis by the host.
|
|
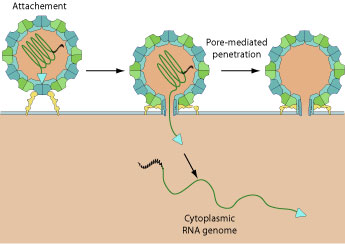
| Binding to receptor leads to a conformational change in capsid structure during which the VP4 is externalized. The hydrophobic N-termini of five VP4 penetrates into host membrane and creates a pore through which the viral genome penetrates into cell cytoplasm.
|

|
Poliovirus virion binds to host Pvr receptor (CD155) via depressions on capsid surface. Virion binding to receptors triggers endocytosis by the host.
|

|
| Binding to receptor leads to a conformational change in capsid structure during which the VP4 is externalized. The hydrophobic N-termini of five VP4 penetrates into host membrane and creates a pore through which the viral genome penetrates into cell cytoplasm.
|
|
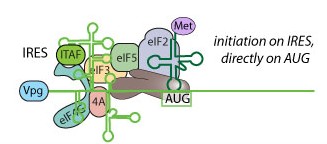
| Poliovirus translation occurs in a cap-independent manner facilitated by the Internal Ribosome Entry Site (IRES). The IRES is a RNA structure in the 5'UTR of the genomic RNA that promotes translation initiation by recruiting initiation factors and help to place the ribosome on the AUG.
|
|
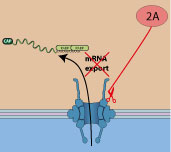
Poliovirus protease 2A cleaves nuclear pore proteins NUP62 and NUP214, impairing host mRNA export out of the nucleus. This action shutoff cellular gene expression.
|
|
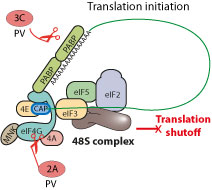
| Poliovirus protease 2A cleaves eIF4G host translation factor in such a way that the translation machinery is still functional for virus translation but insufficient for host mRNA translation. The 3C protease may also play a role in this shutoff.
|
Poliovirus replication proteins localize at membranous vesicles presumably originated from traffic vesicles between endoplasmic reticulum and Golgi apparatus. It is not clear whether the poliovirus replication complex is outside or inside the vesicle. It is represented inside here, because this configuration may help to hide the dsRNA replication intermediate.
The poliovirus genome is translated from a single ORF into a large polyprotein. The first cleavages in cis by 2Apro and 3Cpro produce the P1, P2 and P3 precursors.
These precursors are processed by the 3Cpro into mature peptides. P3 major cleavage produces 3AB and 3CD, minor cleavage 3A, VPg, 3C and 3Dpol.
|
|
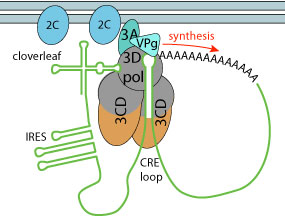
3D polymerase and two 3CD subunits bind to the cre and cloverleaf sequences, inducing adi-uridylylation of VPg protein. The modified VPg is then used as primer to replicate genomic RNA, initiating on the polyA tail. The first round of replication creates a dsRNA which is in turn used for (+) RNA synthesis.
|
(-)genomic RNA is used as template to synthesize (+) RNA genomes. These new genomes can either be encapsidated to join in the cytoplasmic pool of translated genomes, or be encapsidated to form new virions.
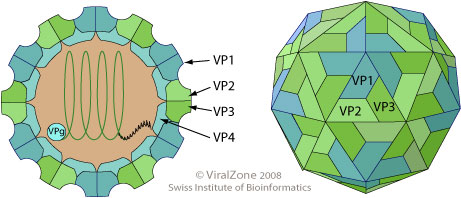
The poliovirus capsid self assembles into a pseudo T=3 icosahedral capsid, composed of VP0, VP2 and VP3. Upon genomic RNA encapsidation the VP0 matures and is cleaved into VP2 and VP4, maybe by self cleavage. This maturation step presumably assures that the membrane pore formation of VP4 is only potentially active within the mature virion, and could prevent early activation in the host cell.
At the end of poliovirus life-cycle the host cell is lysed, releasing new virions.
Enterovirus ViralZone main page
Translation:
The 5' VPg can be cleaved off the genomic RNA by host TBP2, also called "unlinkase"  . This creates a pool genomes with a 5'pUp used for translation/replication. The genomes that are encapsidated directly after synthesis keep the VPg in 5', which may be part of the encapsidation signal.
. This creates a pool genomes with a 5'pUp used for translation/replication. The genomes that are encapsidated directly after synthesis keep the VPg in 5', which may be part of the encapsidation signal.
Replication:
RNA replication is primed by the VPg protein. It would mean that for each replicated RNA, one entire polyprotein has to be synthesized, because there is only one VPg per polyprotein. Host TBP2 releasing the VPg from the genomic RNA may recycle it for more synthesis. Interestingly aphtoviruses encode three subsequent VPg in their polyprotein.
Genome replication occurs in two phases, first the minus strand is synthesized, which is in turn used as template to produce a lot of positive strand RNA genomes. Synthesis of negative strand presumably produces dsRNA. This dsRNA may be the replicative form of picornaviruses. It is possible that the minus strand is separated from the positive strand, but it would require some kind of nucleoprotein to avoid annealing of complementary strand. dsRNA have been detected in replication centers of many positive strand RNA viruses. It may be the template for (+) RNA synthesis by RNA strand displacement.
The dsRNA intermediate may explain why most positive strand RNA viruses replicate in vesicles, that would help to hide the dsRNA from the eukaryotic cell which react very strongly when it detects dsRNA through host PKR or other antiviral sensors.
Encapsidation:
Encapsidation is closely coupled to RNA replication. VPg acts as a replication signal. The new particles seem to assemble at the membranous replication vesicles.
The pseudoT=3 icosahedric capsid requires 60 units of each VP1, VP2 VP3 and VP4 proteins. That represents 60 polyproteins translated to produce a virion, and would mean also 60 polymerases created to get a single virion. This is a bit unusual in virology, with most viruses over-expressing their structural proteins, but express often few polymerases. Some cardioviruses have been shown to use a ribosomal frameshift to translate only the P1 part of their genome  . It is possible that such a mechanism exist in enterovirus for efficient structural proteins translation.
. It is possible that such a mechanism exist in enterovirus for efficient structural proteins translation.









 The poliovirus capsid self assembles into a pseudo T=3 icosahedral capsid, composed of VP0, VP2 and VP3. Upon genomic RNA encapsidation the VP0 matures and is cleaved into VP2 and VP4, maybe by self cleavage. This maturation step presumably assures that the membrane pore formation of VP4 is only potentially active within the mature virion, and could prevent early activation in the host cell.
The poliovirus capsid self assembles into a pseudo T=3 icosahedral capsid, composed of VP0, VP2 and VP3. Upon genomic RNA encapsidation the VP0 matures and is cleaved into VP2 and VP4, maybe by self cleavage. This maturation step presumably assures that the membrane pore formation of VP4 is only potentially active within the mature virion, and could prevent early activation in the host cell.
 . This creates a pool genomes with a 5'pUp used for translation/replication. The genomes that are encapsidated directly after synthesis keep the VPg in 5', which may be part of the encapsidation signal.
. This creates a pool genomes with a 5'pUp used for translation/replication. The genomes that are encapsidated directly after synthesis keep the VPg in 5', which may be part of the encapsidation signal. . It is possible that such a mechanism exist in enterovirus for efficient structural proteins translation.
. It is possible that such a mechanism exist in enterovirus for efficient structural proteins translation.