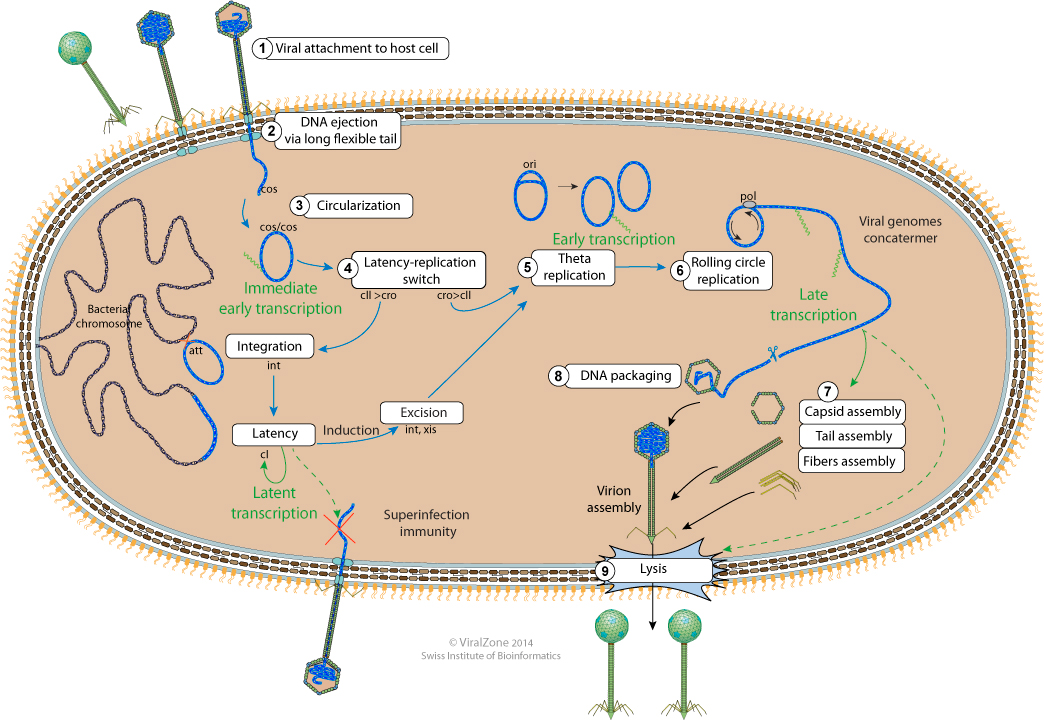
Bacteriophage lambda replication cycle
Lambda fibers attach to host molecules,
until tail tip interacts with entry receptor LamB
until tail tip interacts with entry receptor LamB
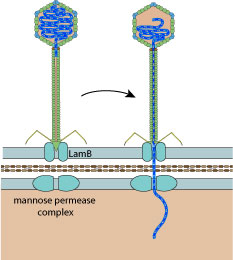 The tail tip interacts with the maltose porin lamB, inducing phage DNA ejection through lamB and the the mannose permease complex in the inner membrane.
The tail tip interacts with the maltose porin lamB, inducing phage DNA ejection through lamB and the the mannose permease complex in the inner membrane.
The cytoplasmic viral DNA circularizes at cos sites and both ends are ligated by the host ligase. Host DNA gyrase puts negative supercoils in the circular genome.
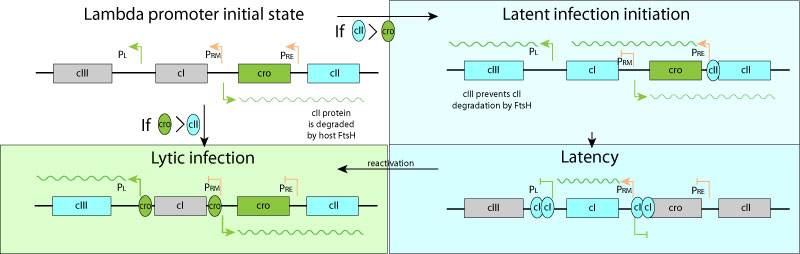 The lambda promoter initially expresses cro and cII proteins, the latter being degraded by host FtsH.
The lambda promoter initially expresses cro and cII proteins, the latter being degraded by host FtsH.
-If cro concentration is higher than cII, the virus enters lytic cycle ans expresses replication program.
-If cII reaches a higher concentration than cro anyway, the genome entres latency. cIII is expressed and protects cII from degradation. When enough cI is produces all promoters are shut off, except Prm expressing cI. At a certain level of cI Prm is shut off.
cII activates the promoter Pi which transcribes the integrase mRNA.Lambda genome is integrated at attB site in bacterial genome.
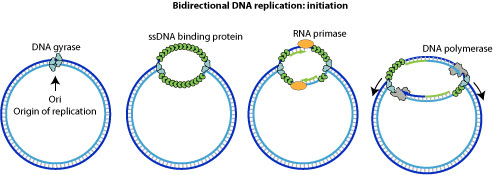 Cellular replication machinery starts replicating the circular virion by bi-directional replication at viral ori site.
Cellular replication machinery starts replicating the circular virion by bi-directional replication at viral ori site.
During latency only cI is expressed, shutting off all viral promoters at high concentration
Upon cellular stress, cI is degraded, reactivating the early left and right viral promoters. Integrase (int) is expressed with exisionase (xis)from the integrated provirus, and together they promote viral genome excision.
Viral replication machinery replicates lambda genome by rolling circle, producing linear genome concatemers.
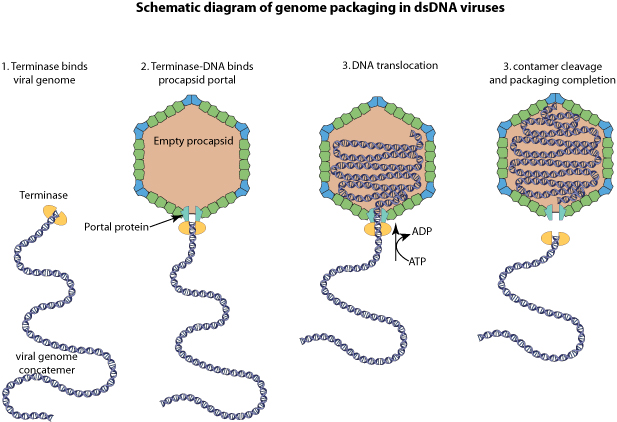 Terminase binds to 5' genome concatemers, and by binds to empty procapsids.
Viral genomic DNA is packaged in an energy dependent manner, and the terminase cleaves the cos site at the end of the genome.
Terminase binds to 5' genome concatemers, and by binds to empty procapsids.
Viral genomic DNA is packaged in an energy dependent manner, and the terminase cleaves the cos site at the end of the genome.
The procapsid is assembled with portal, scaffolding, protease and capsid proteins.
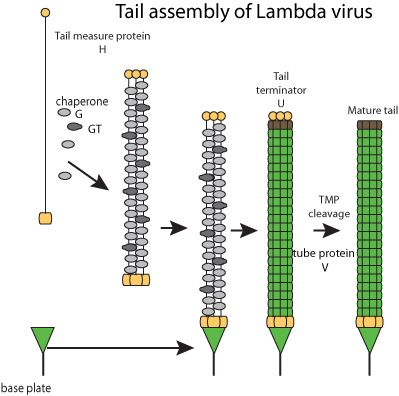
Fibers self-assembles and bind to tail tip