Subgenomic RNA transcription
Subgenomic RNAs of positive-strand viruses
have the same 3' ends as genomic RNA, but have deletions
at the 5' ends to bring the 5' end of the RNA in
proximity with the start codon of downstream (on
genomic RNA) ORFs . Because replication is required for sgRNA synthesis, the RNA-dependent RNA polymerase (RdRp) is always translated first, directly from genomic RNA of positive-strand RNA viruses. sgRNAs express products needed during intermediate and late stages of
infection, such as structural or movement proteins.
These sgRNA are not part of the genome because they don't comprise encapsidation signals and therefore are not carried out by virions to new infected cells.
. Because replication is required for sgRNA synthesis, the RNA-dependent RNA polymerase (RdRp) is always translated first, directly from genomic RNA of positive-strand RNA viruses. sgRNAs express products needed during intermediate and late stages of
infection, such as structural or movement proteins.
These sgRNA are not part of the genome because they don't comprise encapsidation signals and therefore are not carried out by virions to new infected cells.
subgenomic RNA synthesis can be explained two models, it is not clearly established if only one is exact, or if the two happen depending on viruses.
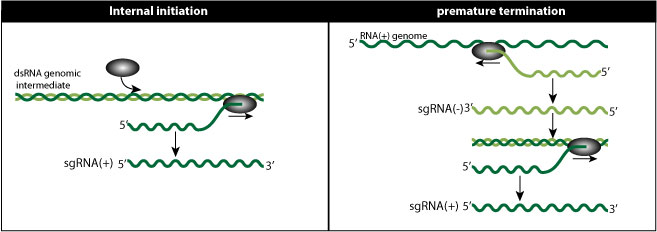
Internal initiation model:
In this model, the viral polymerase somehow initiates not in 3' of genomic RNA, but internally, thereby creating a smaller subgenomic RNA.
Premature termination model (PT model)
The viral polymerase would pause and stop synthesis during minus-RNA synthesis, thereby creating subgenomic minus-RNA shorter that the genomic RNA. This premature termination would be induced by a secondary structure in the genome sequence. The shorter minus-RNA would be template for subgenomic plus-RNAs.
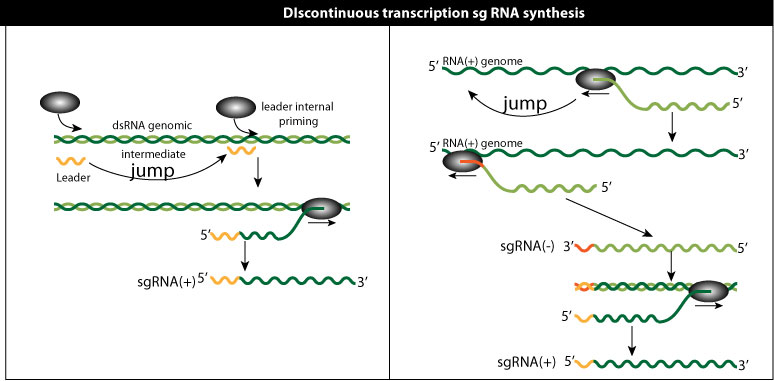
Discontinuous transcription
Discontinuous transcription can happen in the two models above. It's a feature well described in Nidovirales , for which the PT model is favored. Somehow, a leader RNA from the 5' genomic RNA is added to all subgenommic RNA. In internal initiation model it means that leaders RNA are transcribed and serve to prime subgenomic RNAs. In the premature termination model the polymerase would "jump"
, for which the PT model is favored. Somehow, a leader RNA from the 5' genomic RNA is added to all subgenommic RNA. In internal initiation model it means that leaders RNA are transcribed and serve to prime subgenomic RNAs. In the premature termination model the polymerase would "jump"
Alexander O Pasternak, Willy J M Spaan, Eric J Snijder
J. Gen. Virol. June 2006; 87: 1403?1421
W A Miller, G Koev
Virology July 20, 2000; 273: 1-8
Sadaf D Jiwan, K Andrew White
RNA Biol April 2011; 8: 287-294
