Alternative splicing
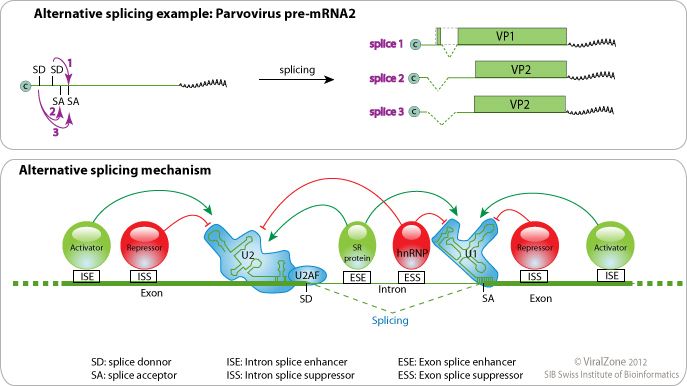
Alternative splicing occurs when several splice donors and/or acceptors are eligible and facultative. This leads to transcripts having different splicing outcome and/or are unspliced. Alternative splicing is regulated by cellular and viral proteins which modulates locally the activity of splicing factors U1 and U2. -It offers the opportunity to encode several proteins in few messengers, like for Adenoviridae and Retroviridae encoding up to 12 different peptides from one pre-mRNA. -It is a way to regulate early and late expression for viruses like Papillomaviridae and maybe Orthomyxoviridae. -Cellular unspliced mRNA cannot be exported out of the nucleus. Hepadnaviridae and Retroviridae have evolved proteins to export their unspliced genomic RNA. -It is used by Herpesviridae as a potential anti-host defense mechanism. By inhibiting some host splicing factors, these viruses prevent the synthesis of key antiviral proteins like PML or STAT1.
| Virus | viral factor | cellular factor | Ref |
| ssDNA viruses | |||
| Anelloviridae | 
| ||
| Circoviridae |  | ||
| Parvoviridae |  | ||
| dsDNA viruses | |||
| Adenoviridae | 33k : expression of late proteins |  | |
| Baculoviridae |  | ||
| Herpesviridae | Unspliced RNA nuclear export EBV- BM2 HCMV- UL69 HSV1- ICP27 |
EBV HCMV
HCMV HSV-1
HSV-1 | |
| Papillomaviridae | E2 activates cellular SR | hnRNP A1 inhibit L1 mRNA expression |   |
| Polyomaviridae | SF2 (SV40) |  | |
| Reverse-transcribing viruses | |||
| Hepadnaviridae | Capsid?: pg-RNA nuclear export |   | |
| Retroviridae, Alpharetrovirus | GAG genomic RNA nuclear export |

| |
| Retroviridae, Lentivirus | genomic RNA nuclear export HIV-1 Rev HTLV-1 Rex | HIV-1 HTLV-1
HTLV-1 | |
| ss(-)RNA viruses | |||
| Bornaviridae |  | ||
| Orthomyxoviridae | NS1 | SF2 |  |
