Viral genome evolution
Viral genomes are the fastest evolving entities in biology, because of their short replication time and the large quantity of offspring released per replication cycle. Evolution occurs via several mechanisms which include: Random mutation, recombination, reassortment, gene amplification/reduction; and results in quasispecies and defective interfering genomes
Random mutation [genetic drift]:
Virus genomes display a higher mutation rate than cellular organisms  . Viral polymerases are more error prone than cellular polymerases, with the least accurate being viral RNA-dependent RNA polymerases
. Viral polymerases are more error prone than cellular polymerases, with the least accurate being viral RNA-dependent RNA polymerases  . Moreover, most RNA viruses or cytoplasmic DNA viruses can't use the cellular proofreading machinery which is functional for nuclear dsDNA, and they generally don't encode this function. The only RNA viruses know to encode for a proofreading are the coronaviridae
. Moreover, most RNA viruses or cytoplasmic DNA viruses can't use the cellular proofreading machinery which is functional for nuclear dsDNA, and they generally don't encode this function. The only RNA viruses know to encode for a proofreading are the coronaviridae  .
Error prone polymerases and no proofreading are responsible for a high viral genome mutation rate. This produces mainly deficient genomes, but this drawback is counteracted by the very high number of progeny produced per virion
.
Error prone polymerases and no proofreading are responsible for a high viral genome mutation rate. This produces mainly deficient genomes, but this drawback is counteracted by the very high number of progeny produced per virion 
 .
.
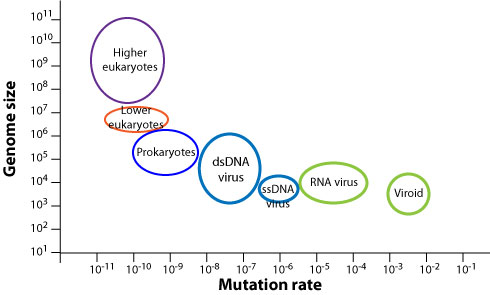
The mutation rate represents the number of mutations per site per replication  . Mutations can be deleterious, neutral or beneficial. Deleterious mutations are quickly lost. Neutral and beneficial mutations circulate in a viral population and can be observed by genetic surveillance. Beneficial mutations tend to be fixed by natural selection. The rate of observed genomic evolution is measured by the substitution rate (also called evolution rate). It is much lower than the mutation rate because deleterious mutations are lost, and it varies between viruses
. Mutations can be deleterious, neutral or beneficial. Deleterious mutations are quickly lost. Neutral and beneficial mutations circulate in a viral population and can be observed by genetic surveillance. Beneficial mutations tend to be fixed by natural selection. The rate of observed genomic evolution is measured by the substitution rate (also called evolution rate). It is much lower than the mutation rate because deleterious mutations are lost, and it varies between viruses 
 .
.
Recombination by crossing over [genetic shift]:
Genetic recombination is the exchange of part of genome between two genetic entities. There are two kinds of recombination: self-recombination and recombination with host.
Self recombination occurs when two closely related viral genomes recombine by homologous crossing over. This happens for DNA viruses  and is very common in prokaryotic viruses
and is very common in prokaryotic viruses . Some RNA viruses also evolve by recombination
. Some RNA viruses also evolve by recombination 
 , it is common for retroviruses
, it is common for retroviruses  . These recombination events can be an evolutionary advantage for the virus when they help it evade host immune defenses, for example by changing surface protein antigenicity.
Recombination with host or other organism occurs when a viral genome acquires sequences from a cellular organism
. These recombination events can be an evolutionary advantage for the virus when they help it evade host immune defenses, for example by changing surface protein antigenicity.
Recombination with host or other organism occurs when a viral genome acquires sequences from a cellular organism  . This allows the virus to capture cellular genes and highjack their function, which can provide an evolutionary advantage for the virus. This event is common in large dsDNA viruses like poxviridae, and it is used in biotechnology to produce vaccines
. This allows the virus to capture cellular genes and highjack their function, which can provide an evolutionary advantage for the virus. This event is common in large dsDNA viruses like poxviridae, and it is used in biotechnology to produce vaccines  . Protist and algual nuclear-cytoplasmic large DNA viruses (NCLDV) may have acquired many genes from bacteria that are prey of the eukaryotic host cell
. Protist and algual nuclear-cytoplasmic large DNA viruses (NCLDV) may have acquired many genes from bacteria that are prey of the eukaryotic host cell  .
.
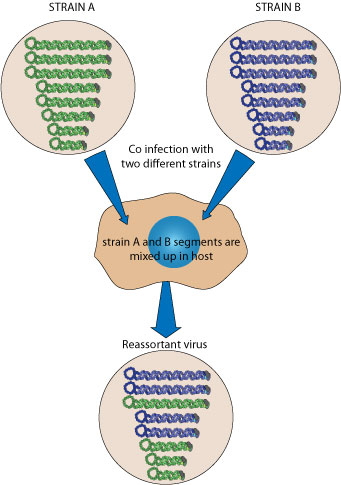
Segment reassortment [genetic shift]:
Reassortment occurs event when two similar segmented viruses exchange part of their genomes during a cell co-infection 


 . This way segmented viruses can modify quickly their antigens, creating a new strain that can evade the previously established immune protection of a population. This has been particularly well studied for influenza virus, since reassortment is the major event which gives rise to new flu pandemics
. This way segmented viruses can modify quickly their antigens, creating a new strain that can evade the previously established immune protection of a population. This has been particularly well studied for influenza virus, since reassortment is the major event which gives rise to new flu pandemics .
.
Gene amplification/reduction
Gene amplification is a process that produces extra copies of the same gene in a viral genome. In poxviridae, host genes that have been included in the viral genome by recombination can be subject to amplification. The host gene will be multiplicated on the viral genome. More copies increase the probability of beneficial mutations  . After acquisition of a beneficial mutation, these viruses can lose the non-beneficial copies.
. After acquisition of a beneficial mutation, these viruses can lose the non-beneficial copies.
Consequences of viral evolution:
-Viral quasispecies: The fast and flexible evolution of RNA virus genomes creates population of viruses with a large numbers of variant genomes. A single sequence cannot accurately describe the viral population in a single host or even in cell culture  . This provides an evolutionary advantage, because mutations are already present and waiting to be selected, when selection pressure is applied to such a population.
-Defective interfering virus:
Defective genomes can arise through deletion, ?rearrangement, or recombination of a competent viral genome
. This provides an evolutionary advantage, because mutations are already present and waiting to be selected, when selection pressure is applied to such a population.
-Defective interfering virus:
Defective genomes can arise through deletion, ?rearrangement, or recombination of a competent viral genome  . In negative stranded RNA viruses, these are called defective interfering viruses (DI) which compete with the viral genome for replication and/or encapsidation: this tends to attenuate the virus and triggers host antiviral defenses. These defective genomes accumulate in cell culture, where innate antiviral defense is often lacking. For example, Sendai virus grown in cell culture is full of DIs, which are very potent inducers of innate immunity. Indeed DIs are not encoding for viral genes that downregulate host interferon system. Therefore, the more DI there is with the virus, the less the virus will be able to tone down interferon induction. That is why cultured Sendai virus is commonly used to experimentally induce interferon, whereas the competent Sendai virus would efficiently shut off the IFN production.
. In negative stranded RNA viruses, these are called defective interfering viruses (DI) which compete with the viral genome for replication and/or encapsidation: this tends to attenuate the virus and triggers host antiviral defenses. These defective genomes accumulate in cell culture, where innate antiviral defense is often lacking. For example, Sendai virus grown in cell culture is full of DIs, which are very potent inducers of innate immunity. Indeed DIs are not encoding for viral genes that downregulate host interferon system. Therefore, the more DI there is with the virus, the less the virus will be able to tone down interferon induction. That is why cultured Sendai virus is commonly used to experimentally induce interferon, whereas the competent Sendai virus would efficiently shut off the IFN production.
A C Marriott, N J Dimmock
Rev. Med. Virol. January 2010; 20: 51-62
Siobain Duffy, Laura A Shackelton, Edward C Holmes
Nat. Rev. Genet. April 2008; 9: 267-276
Joanna Sztuba-Soliska, Anna Urbanowicz, Marek Figlerowicz, Jozef J Bujarski
Annu Rev Phytopathol 2011; 49: 415-443
Adewunmi Onafuwa-Nuga, Alice Telesnitsky
Microbiol. Mol. Biol. Rev. September 2009; 73: 451-480, Table of Contents
A N Lukashev
Rev. Med. Virol. September 2010; 20: 327-337
Darren P Martin, Philippe Biagini, Pierre Lefeuvre, Michael Golden, Philippe Roumagnac, Arvind Varsani
Viruses September 2011; 3: 1699-1738
Matthew L Paff, Steven P Stolte, James J Bull
Mol. Biol. Evol. January 2014; 31: 96-105
Matthew L Paff, Steven P Stolte, James J Bull
Mol. Biol. Evol. January 2014; 31: 96-105
Adam S Lauring, Judith Frydman, Raul Andino
Nat. Rev. Microbiol. May 2013; 11: 327-336
Robert Belshaw, Rafael Sanjun, Oliver G Pybus
Curr Opin Virol November 2011; 1: 430-435
Jonathan File
Curr Opin Virol October 2013; 3: 595-599
Jonathan File, Noelle Pouget, Mick Chandler
BMC Evol. Biol. 2008; 8: 320
John R Roth, D I Andersson
Cell August 17, 2012; 150: 671-672
Thomas Briese, Charles H Calisher, Stephen Higgs
Virology November 2013; 446: 207?216
Rafael Sanju?n
PLoS Pathog. 2012; 8: e1002685
J W Drake
Ann. N. Y. Acad. Sci. May 18, 1999; 870: 100?107
Nicolle Marshall, Lalita Priyamvada, Zachary Ende, John Steel, Anice C Lowen
PLoS Pathog. 2013; 9: e1003421
Hung J Liu, Long H Lee, Hsiao W Hsu, Liam C Kuo, Ming H Liao
Virology September 15, 2003; 314: 336?349
Francisco M Codo?er, Santiago F Elena
J. Gen. Virol. July 2008; 89: 1739?1747
Dhanasekaran Vijaykrishna, Reshmi Mukerji, Gavin J. D. Smith
PLoS Pathog. July 2015; 11: e1004902
Robson F, Khan KS, Le TK, Paris C, Demirbag S, Barfuss P, Rocchi P, Ng WL.Mol Cell. 2020 Sep 3;79(5):710-727.
Duffy S. PLoS Biol. 2018 Aug 13;16(8)
Nafissi N, Slavcev R.
Appl Microbiol Biotechnol. 2014 Apr;98(7):2841-51. doi: 10.1007/s00253-014-5512-2. Epub 2014 Jan 19.
Henderson DA, Moss B.
In: Plotkin SA, Orenstein WA, editors. Vaccines. 3rd edition.
Curr Top Microbiol Immunol. 2016
