DNA end degradation evasion by virus (kw:KW-1256)
Several types of bacteria including Escherichia coli or Salmonella typhimurium encode enzymes responsible for the degradation of free DNA ends. When functioning in their exonuclease mode, helicase/nuclease proteins such as bacterial RecBCD (also called exonuclease V) or AddAB degrade any free DNA ends as well as linear dsDNA products resulting from restriction Type II cleavage  .
.
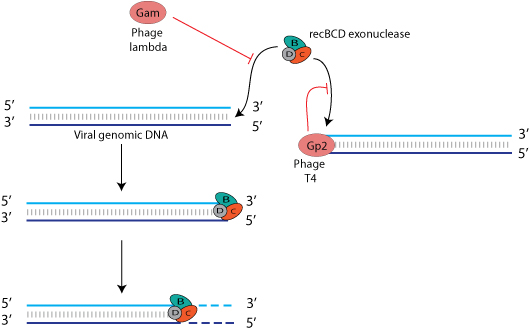
Any bacterial virus that exposes free DNA ends as part of its life cycle must find a means to evade destruction by these host nucleases. Bacterial viruses have elaborated different strategies to circumvent degradation of their free DNA ends. For example, bacteriophage T4 gene product 2 (gp2) is able to interact with viral DNA ends to prevent recognition of the RecBCD complex and subsequent cleavage of the DNA. Gam protein of bacteriophage lambda also inhibits the interaction between RecBCD and viral genome ends.
Mark S Dillingham, Stephen C Kowalczykowski
Microbiol. Mol. Biol. Rev. December 2008; 72: 642-671, Table of Contents
B Lipinska, A S Rao, B M Bolten, R Balakrishnan, E B Goldberg
J. Bacteriol. January 1989; 171: 488-497
Kenan C Murphy
J. Mol. Biol. August 3, 2007; 371: 19-24
d'Adda di Fagagna F, Weller GR, Doherty AJ, Jackson SP
EMBO Rep. 2003 Jan;4(1):47-52
Robert Court, Nicola Cook, Kayarat Saikrishnan, Dale Wigley
J. Mol. Biol. August 3, 2007; 371: 25-33
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)6 entries grouped by protein
1 entry
DNA circularization protein N (64 kDa virion protein) (Gene product 43) (gp43) (Gene product N) (gpN)
1 entry
Terminal DNA protecting protein (Gene product 64) (gp64) (Head protein Gp2)
2 entries
Host-nuclease inhibitor protein gam
2 entries
