Ascoviridae (taxid:43682)
VIRION
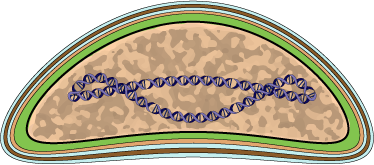
Virions consist of an envelope, a core, and an internal lipid membrane associated with the inner particle. Virion can be bacilliform, ovoidal or allantoid in shape, mesuring about 130 nm in diameter, by 200-400 nm in length. Contains at least 15 different proteins.
GENOME
Circular, dsDNA genome of 156-186 kb. The genome contains two tandem of inverted repeats.
GENE EXPRESSION
Encode for up to 180 open reading frames.
ENZYMES
- DNA-directed DNA polymerase [ORF01]
- DNA-directed RNA polymerase [ORF052]
- Protease (Peptidase C14) [ORF052]
- Protease (Peptidase M10) [ORF14]
REPLICATION
NUCLEUS
- Attachment of the viral proteins to host receptors mediates endocytosis of the virus into the host cell.
- Fusion with the plasma membrane occurs; and the viral DNA is released into the nucleus.
- Transcription of viral genes, replication of the DNA genome in the nucleus.
- Cell nucleus enlarges and ruptures.
- Assembly of new virions in the cytoplasm.
- Host cell is cleaved in cluster of virion-containing vesicles.
Host-virus interaction
Apoptosis modulation
The Spodoptera frugiperda ascovirus encodes a viral caspase that is able to induce apoptosis in insect cells.
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)34 entries grouped by strain
12 entries
Heliothis virescens ascovirus 3e (HvAV-3e) reference strain
11 entries
Spodoptera frugiperda ascovirus 1a (SfAV-1a) reference strain
11 entries
