Eukaryotic host translation shutoff by virus (kw:KW-1193)
The majority of cellular mRNAs initiate translation through the recruitment of a multisubunit translation initiation complex termed eIF4F, which consists of the cap-binding protein eIF4E, the RNA helicase eIF4A, and the adaptor protein eIF4G. eIF4G binds poly(A)-binding protein (PABP) to mediate 5' -> 3' communication, probably to promote efficient translation of intact correctly processed mRNAs.
eIF4E binding protein 1 (eIF4E-BP1) functions as a translational repressor that limits eIF4E availability and therefore eIF4F complex formation.
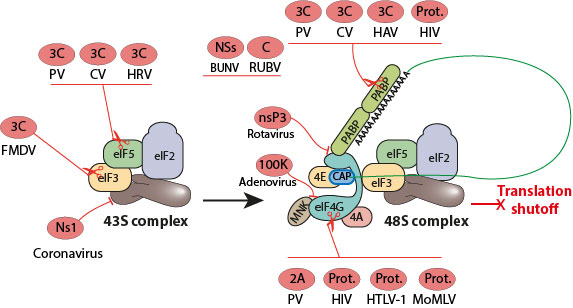
Viruses have evolved ways of interacting with the host translational machinery to shutoff host gene expression. This global inhibition of cellular protein synthesis serves to ensure maximal viral gene expression and to evade host immune response

 .
.
All the viruses inducing the shutoff of translation are able to continue to translate at least part of their mRNAs using non-canonical translation: IRES, Ribosome shunting, or VPG initiation. For adenoviridae, polyomaviridae and togaviridae the cellular translation shutoff takes place at the late phase of infection and ensures an optimal production of viral structural proteins.
Under stress conditions such as viral infection some cellular translation can continue when the cap-dependent translation initiation is inhibited. Expression of specific cellular proteins seems to occur through cap-independent mechanisms

 .
.
Viruses inducing host translation shutoff:
| Family | Virus | Viral protein | Shutoff strategy | Viral translation | ref. |
| Adenoviridae | Adenovirus | Shutoff protein 100K | Inhibits eIF4G-Mnk1 and prevents EIF4E phosphorylation | Ribosome shunting on late mRNAs |


|
| Bunyaviridae | Bunyamwera virus | Nucleoprotein | Nuclear retention of PABP | Viral mRNAs non polyadenylated | 
|
| Caliciviridae | Norovirus Feline calicivirus | Protease 3C Protease |
PABP cleavage | VPg
 |

|
| Feline calicivirus | Protease | Cleavage of eIF4G1 and eIF4G2 | VPg  |
 |
|
| Dicistroviridae | Cripavirus | ? | Dissociation of eIF4G and eIF4E | IRES |

|
| Orthomyxoviridae | Influenza virus | ? | Dephosphorylation of of eIF-4E | eIF-4E independent translation of viral mRNAs |


|
| Picornaviridae | Poliovirus | Protease 2A | Cleavage of eIF4GI and eIF4GII | IRES |


|
| Protease 3C | Cleavage of eIF5B | IRES |

|
||
| Protease 3C | PABP cleavage | IRES |

|
||
| Rhinovirus | Protease 2A | Cleavage of eIF4GI and eIF4GII | IRES |


|
|
| Coxsackie virus | Protease 2A | Cleavage of eIF4GI and eIF4GII | IRES |


|
|
| Coxsackie virus | Protease 2A,3C | PABP cleavage | IRES |


|
|
| Protease 3C | Cleavage of eIF5B | IRES |

|
||
| Encephalomyocarditis virus | Protein 2A | ? | IRES |

|
|
| ? | Dephosphorylation of 4E-BP1 | IRES |


|
||
| Foot-and-mouth disease virus | Leader protease | Cleavage of eIF4GI and eIF4GII | IRES |

 |
|
| Protease 3C | Cleavage of PABP, eIF3a, eIF3b, and eIF4A | IRES |

|
||
| Polyomaviridae | SV40 | Small T antigen | Dephosphorylation of 4E-BP1 | IRES |


|
| Reoviridae | Rotavirus | NSP3 | Interacts with EIF4G and evicts PABP from initiation complexes. Nuclear relocalization of PABP | NSP3 replaces PABP and bind the non polyAdenylated viral mRNAs |


|
| Rhabdoviridae | Vesicular stomatitis virus | ? | Dephosphorylation of 4E-BP1 | classical |

|
| Retroviridae | HIV-1 | Protease | Cleavage of eIF4GI and PABP | IRES |



|
| HTLV-1 | Protease | Cleavage of eIF4GI | IRES |

|
|
| Moloney murine leukemia virus | Protease | Cleavage of eIF4GI and eIF4GII | IRES |

|
|
| Togaviridae | Rubella virus | Capsid protein | PABP sequestration | classical |

|
| Sindbis virus | dsRNA leading to PKR-mediated inhibition of eIF2alpha | DLP |


| ||
| Coronaviridae | SARS-CoV | Nsp1 | Binding to host 40S subunit | Viral mRNA protected by 5'-end leader sequence |


|
| Bat-CoV | Nsp1 | - | - |

|
|
| TGEV | Nsp1 | - | - |

|
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)203 entries grouped by strain
2 entries
Bat coronavirus HKU4 (BtCoV) (BtCoV/HKU4/2004) reference strain
2 entries
Bat coronavirus HKU5 (BtCoV) (BtCoV/HKU5/2004) reference strain
2 entries
Bat coronavirus HKU9 (BtCoV) (BtCoV/HKU9) reference strain
2 entries
Human T-cell leukemia virus 1 (isolate Caribbea HS-35 subtype A) (HTLV-1) reference strain
2 entries
Human T-cell leukemia virus 2 (HTLV-2) reference strain
2 entries
Human T-cell leukemia virus 3 (strain 2026ND) (HTLV-3) reference strain
2 entries
Human coronavirus HKU1 (isolate N1) (HCoV-HKU1) reference strain
2 entries
Human coronavirus OC43 (HCoV-OC43) reference strain
2 entries
Murine coronavirus (strain A59) (MHV-A59) (Murine hepatitis virus) reference strain
2 entries
Severe acute respiratory syndrome coronavirus (SARS-CoV) reference strain
2 entries
Severe acute respiratory syndrome coronavirus 2 (2019-nCoV) (SARS-CoV-2) reference strain
1 entry
African swine fever virus (strain Badajoz 1971 Vero-adapted) (Ba71V) (ASFV) reference strain
1 entry
Aichi virus (strain Human/A846/88/1989) (AiV) (Aichi virus (strain A846/88)) reference strain
1 entry
Bovine enterovirus (strain VG-5-27) (BEV) reference strain
1 entry
Bunyamwera virus (BUNV) reference strain
1 entry
Bunyavirus La Crosse (isolate Human/United States/L78/1978) reference strain
1 entry
Canine adenovirus serotype 1 (strain RI261) (CAdV-1) (Canine adenovirus 1 (strain RI261)) reference strain
1 entry
Encephalomyocarditis virus (strain Rueckert) (EMCV) reference strain
1 entry
Feline calicivirus (strain Cat/United States/Urbana/1960) (FCV) reference strain
1 entry
Feline calicivirus (strain F9) (FCV) reference strain
1 entry
Fowl adenovirus A serotype 1 (strain CELO / Phelps) (FAdV-1) (Avian adenovirus gal1 (strain Phelps)) reference strain
1 entry
Human adenovirus A serotype 12 (HAdV-12) (Human adenovirus 12) reference strain
1 entry
Human adenovirus C serotype 2 (HAdV-2) (Human adenovirus 2) reference strain
1 entry
Human adenovirus F serotype 40 (HAdV-40) (Human adenovirus 40) reference strain
1 entry
Human enterovirus D68 (EV68) (EV-68) reference strain
1 entry
Human immunodeficiency virus type 1 group M subtype A (isolate U455) (HIV-1) reference strain
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate HXB2) (HIV-1) reference strain
1 entry
Human immunodeficiency virus type 1 group N (isolate YBF30) (HIV-1) reference strain
1 entry
Human immunodeficiency virus type 1 group O (isolate ANT70) (HIV-1) reference strain
1 entry
Human immunodeficiency virus type 2 subtype A (isolate BEN) (HIV-2) reference strain
1 entry
Human rhinovirus 1A (HRV-1A) reference strain
1 entry
Human rhinovirus 3 (HRV-3) reference strain
1 entry
Human rhinovirus A serotype 89 (strain 41467-Gallo) (HRV-89) reference strain
1 entry
Moloney murine leukemia virus (isolate Shinnick) (MoMLV) reference strain
1 entry
Monkeypox virus (MPXV) reference strain
1 entry
Poliovirus type 1 (strain Mahoney) reference strain
1 entry
Porcine adenovirus A serotype 3 (PAdV-3) (Porcine adenovirus 3) reference strain
1 entry
Salivirus A (isolate Human/Nigeria/NG-J1/2007) (SV-A) reference strain
1 entry
Simian immunodeficiency virus agm.grivet (isolate AGM gr-1) (SIV-agm.gri) (Simian immunodeficiency virus African green monkey grivet) reference strain
1 entry
Simian virus 40 (SV40) reference strain
1 entry
Snake adenovirus serotype 1 (SnAdV-1) reference strain
1 entry
Vaccinia virus (strain Western Reserve) (VACV) (Vaccinia virus (strain WR)) reference strain
2 entries
Bat coronavirus 133/2005 (BtCoV) (BtCoV/133/2005)
2 entries
Bat coronavirus 279/2005 (BtCoV) (BtCoV/279/2005)
2 entries
Bat coronavirus HKU3 (BtCoV) (SARS-like coronavirus HKU3)
2 entries
Bat coronavirus Rp3/2004 (BtCoV/Rp3/2004) (SARS-like coronavirus Rp3)
2 entries
Bovine coronavirus (strain 98TXSF-110-ENT) (BCoV-ENT) (BCV)
2 entries
Bovine coronavirus (strain 98TXSF-110-LUN) (BCoV-LUN) (BCV)
2 entries
Bovine coronavirus (strain Mebus) (BCoV) (BCV)
2 entries
Bovine coronavirus (strain Quebec) (BCoV) (BCV)
2 entries
Human T-cell leukemia virus 1 (isolate Melanesia mel5 subtype C) (HTLV-1)
2 entries
Human T-cell leukemia virus 1 (strain Japan ATK-1 subtype A) (HTLV-1)
2 entries
Human T-cell leukemia virus 3 (strain Pyl43) (HTLV-3)
2 entries
Human coronavirus HKU1 (isolate N2) (HCoV-HKU1)
2 entries
Human coronavirus HKU1 (isolate N5) (HCoV-HKU1)
2 entries
Middle East respiratory syndrome-related coronavirus (isolate United Kingdom/H123990006/2012) (MERS-CoV) (Betacoronavirus England 1)
2 entries
Murine coronavirus (strain 2) (MHV-2) (Murine hepatitis virus)
2 entries
Murine coronavirus (strain JHM) (MHV-JHM) (Murine hepatitis virus)
1 entry
AKV murine leukemia virus (AKR (endogenous) murine leukemia virus)
1 entry
African swine fever virus (isolate Tick/South Africa/Pretoriuskop Pr4/1996) (ASFV)
1 entry
Bunyavirus La Crosse
1 entry
Bunyavirus germiston
1 entry
Bunyavirus snowshoe hare
1 entry
Canine adenovirus serotype 1 (strain CLL) (CAdV-1) (Canine adenovirus 1 (strain CLL))
1 entry
Cas-Br-E murine leukemia virus
1 entry
Coxsackievirus A16 (strain G-10)
1 entry
Coxsackievirus A16 (strain Tainan/5079/98)
1 entry
Coxsackievirus A21 (strain Coe)
1 entry
Coxsackievirus A24 (strain EH24/70)
1 entry
Coxsackievirus A9 (strain Griggs)
1 entry
Coxsackievirus B1 (strain Japan)
1 entry
Coxsackievirus B2 (strain Ohio-1)
1 entry
Coxsackievirus B3 (strain Nancy)
1 entry
Coxsackievirus B3 (strain Woodruff)
1 entry
Coxsackievirus B4 (strain E2) (CVB4)
1 entry
Coxsackievirus B4 (strain JVB / Benschoten / New York/51)
1 entry
Coxsackievirus B5 (strain Peterborough / 1954/UK/85)
1 entry
Coxsackievirus B6 (strain Schmitt)
1 entry
Echovirus 1 (strain Human/Egypt/Farouk/1951) (E-1)
1 entry
Echovirus 11 (strain Gregory)
1 entry
Echovirus 12 (strain Travis)
1 entry
Echovirus 30 (strain Bastianni)
1 entry
Echovirus 5 (strain Noyce)
1 entry
Echovirus 6 (strain Charles)
1 entry
Echovirus 9 (strain Barty)
1 entry
Echovirus 9 (strain Hill)
1 entry
Encephalomyocarditis virus
1 entry
Encephalomyocarditis virus (strain emc-b nondiabetogenic)
1 entry
Encephalomyocarditis virus (strain emc-d diabetogenic)
1 entry
Feline calicivirus (strain CFI/68 FIV) (FCV)
1 entry
Feline calicivirus (strain Japanese F4) (FCV)
1 entry
Fowl adenovirus C serotype 10 (strain SA2) (FAdV-10) (Fowl adenovirus 10)
1 entry
Friend murine leukemia virus (isolate 57) (FrMLV)
1 entry
Friend murine leukemia virus (isolate FB29) (FrMLV)
1 entry
Friend murine leukemia virus (isolate PVC-211) (FrMLV)
1 entry
Human adenovirus C serotype 5 (HAdV-5) (Human adenovirus 5)
1 entry
Human adenovirus F serotype 41 (HAdV-41) (Human adenovirus 41)
1 entry
Human enterovirus 70 (strain J670/71) (EV70) (EV-70)
1 entry
Human enterovirus 71 (EV71) (EV-71)
1 entry
Human enterovirus 71 (strain 7423/MS/87) (EV71) (EV-71)
1 entry
Human enterovirus 71 (strain USA/BrCr/1970) (EV71) (EV-71)
1 entry
Human immunodeficiency virus type 1 group M subtype A (isolate MAL) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate ARV2/SF2) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate BH10) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate BH5) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate BRU/LAI) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate JRCSF) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate LW123) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate MN) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate NY5) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate OYI) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate RF/HAT3) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (isolate YU-2) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype B (strain 89.6) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype C (isolate 92BR025) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype C (isolate ETH2220) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype D (isolate ELI) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype D (isolate NDK) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype D (isolate Z2/CDC-Z34) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype F1 (isolate 93BR020) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype F1 (isolate VI850) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype F2 (isolate MP255) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype F2 (isolate MP257) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype G (isolate 92NG083) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype G (isolate SE6165) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype H (isolate 90CF056) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype H (isolate VI991) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype J (isolate SE9173) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype J (isolate SE9280) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype K (isolate 96CM-MP535) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group M subtype K (isolate 97ZR-EQTB11) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group N (isolate YBF106) (HIV-1)
1 entry
Human immunodeficiency virus type 1 group O (isolate MVP5180) (HIV-1)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate CAM2) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate D194) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate Ghana-1) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate KR) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate NIH-Z) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate ROD) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate SBLISY) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype A (isolate ST) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype B (isolate D205) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype B (isolate EHO) (HIV-2)
1 entry
Human immunodeficiency virus type 2 subtype B (isolate UC1) (HIV-2)
1 entry
Human klassevirus 1 (HKV-1)
1 entry
Human rhinovirus 14 (HRV-14)
1 entry
Human rhinovirus 16 (HRV-16)
1 entry
Human rhinovirus 1B (HRV-1B)
1 entry
Human rhinovirus 2 (HRV-2)
1 entry
Human rhinovirus C (strain C15) (HRV-C15)
1 entry
Mengo encephalomyocarditis virus
1 entry
Murine leukemia virus (strain BM5 eco)
1 entry
Poliovirus type 1 (strain Sabin)
1 entry
Poliovirus type 2 (strain Lansing)
1 entry
Poliovirus type 2 (strain W-2)
1 entry
Poliovirus type 3 (strain 23127)
1 entry
Poliovirus type 3 (strains P3/Leon/37 and P3/Leon 12A[1]B)
1 entry
Porcine enterovirus 9 (strain UKG/410/73)
1 entry
Radiation murine leukemia virus
1 entry
Simian immunodeficiency virus (isolate CPZ GAB1) (SIV-cpz) (Chimpanzee immunodeficiency virus)
1 entry
Simian immunodeficiency virus (isolate EK505) (SIV-cpz) (Chimpanzee immunodeficiency virus)
1 entry
Simian immunodeficiency virus (isolate F236/smH4) (SIV-sm) (Simian immunodeficiency virus sooty mangabey monkey)
1 entry
Simian immunodeficiency virus (isolate GB1) (SIV-mnd) (Simian immunodeficiency virus mandrill)
1 entry
Simian immunodeficiency virus (isolate MB66) (SIV-cpz) (Chimpanzee immunodeficiency virus)
1 entry
Simian immunodeficiency virus (isolate Mm142-83) (SIV-mac) (Simian immunodeficiency virus rhesus monkey)
1 entry
Simian immunodeficiency virus (isolate Mm251) (SIV-mac) (Simian immunodeficiency virus rhesus monkey)
1 entry
Simian immunodeficiency virus (isolate PBj14/BCL-3) (SIV-sm) (Simian immunodeficiency virus sooty mangabey monkey)
1 entry
Simian immunodeficiency virus (isolate TAN1) (SIV-cpz) (Chimpanzee immunodeficiency virus)
1 entry
Simian immunodeficiency virus agm.vervet (isolate AGM TYO-1) (SIV-agm.ver) (Simian immunodeficiency virus African green monkey vervet)
1 entry
Simian immunodeficiency virus agm.vervet (isolate AGM155) (SIV-agm.ver) (Simian immunodeficiency virus African green monkey vervet)
1 entry
Simian immunodeficiency virus agm.vervet (isolate AGM3) (SIV-agm.ver) (Simian immunodeficiency virus African green monkey vervet)
1 entry
Swine vesicular disease virus (strain H/3 '76) (SVDV)
1 entry
Swine vesicular disease virus (strain UKG/27/72) (SVDV)
1 entry
Theiler's murine encephalomyelitis virus (strain BeAn 8386) (TMEV)
1 entry
Theiler's murine encephalomyelitis virus (strain DA) (TMEV)
1 entry
