Batravirus (taxid:3044671)
VIRION
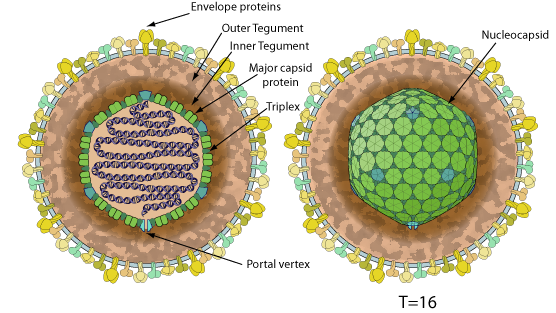
Enveloped, spherical to pleomorphic, 150-200 nm in diameter, T=16 icosahedral symmetry. Capsid consists of 162 capsomers. Glycoproteins complexes are embed in the lipid envelope.
GENOME
Monopartite, linear, extensively methylated dsDNA genome of 220-231 kb. The genome contains terminal and internal reiterated sequences.
GENE EXPRESSION
Each viral transcript usually encodes a single protein and has a promoter/regulatory sequence, a TATA box, a transcription initiation site, a 5' leader sequence of 30-300 bp (not translated), a 3' non-translated sequence of 10-30 bp, and a poly A signal. There are only few spliced genes. Some of the expressed ORFs are antisense to each other. The three exons of the gene encoding the putative ATPase subunit of terminase are not specified by the same DNA strand.
ENZYMES
- DNA-dependent DNA polymerase family B
- Assemblin (Peptidase S8)
- Kinase
- Helicase
- Thymidine kinase
REPLICATION
NUCLEAR
Lytic replication:
- Attachment of the viral glycoproteins to host receptors mediates endocytosis of the virus into the host cell.
- Entry into host cell is still unclear and may depend on the host cell type, i.e. endocytosis versus fusion at the plasma membrane.
- The capsid is transported to the nuclear pore where the viral DNA is released into the nucleus.
- Transcription of immediate early genes which promote transcription of early genes.
- Transcription of early viral mRNA by host polymerase II, transport into the cytoplasm and translation into early proteins.
- Early proteins are involved in replication of the viral DNA and are transported back into the nucleus.
- Synthesis of multiple copies of viral DNA by the viral DNA-dependent DNA polymerase.
- Transcription of late mRNAs by host polymerase II, transport into the cytoplasm and translation into late proteins.
- Late proteins are structural or core proteins and are transported back into the nucleus.
- Assembly of the virus in nuclear viral factories and budding through the inner lamella of the nuclear membrane which has been modified by the insertion of herpes glycoproteins, throughout the Golgi and final release at the plasma membrane.
Latent replication : replication of circular viral episome in tandem with the host cell DNA using the host cell replication machinery.
