Cytomegalovirus (taxid:10358)
VIRION
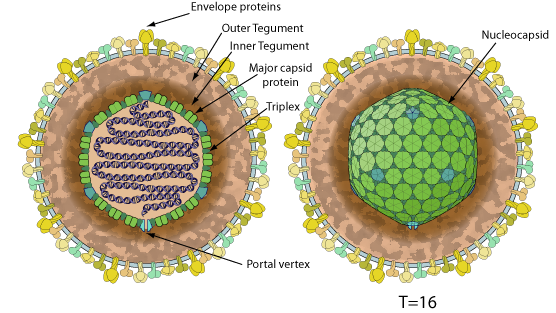
Enveloped, spherical to pleomorphic, 150-200 nm in diameter, T=16 icosahedral symmetry. Capsid consists of 162 capsomers and is surrounded by an amorphous tegument. Glycoproteins complexes are embedded in the lipid envelope.
GENOME
Monopartite, linear, dsDNA genome of about 200 kb. The genome contains terminal and internal reiterated sequences.
GENE EXPRESSION
Each viral transcript usually encodes a single protein and has a promoter/regulatory sequence, a TATA box, a transcription initiation site, a 5' leader sequence of 30-300 bp (not translated), a 3' untranslated sequence of 10-30 bp, and a poly A signal. There are many gene overlaps. There are only few spliced genes. Some of the expressed ORFs are antisense to each other. Some ORFs can be accessed from more than one promoter. Certain proteins are downregulated translationaly by a leaky scanning from an upstream ORF.
ENZYMES
- DNA-dependent DNA polymerase
- DNA primase
- Tegument deneddylase (Peptidase C76)
- Assemblin (Peptidase S21)
- Kinase
- Helicase
- Ribonucleoside-diphosphate reductase
- Uracil-DNA glycosylase
REPLICATION
NUCLEAR
Lytic replication:
- Attachement of the viral glycoproteins to host receptors mediates endocytosis of the virus into the host cell.
- Entry into host cell is still unclear and may depend on the host cell type, i.e. endocytosis versus fusion at the plasma membrane.
- The capsid is transported to the nuclear pore where the viral DNA is released into the nucleus.
- Transcription of immediate early genes which promote transcription of early genes and protect the virus against innate host immunity.
- Transcription of early viral mRNA by host polymerase II, encoding proteins involved in replication of the viral DNA.
- A first round of circular genome amplification occurs by bidirectional replication
- Synthesis of linear concatemer copies of viral DNA by rolling circle.
- Transcription of late mRNAs by host polymerase II, encoding structural proteins.
- Assembly of the virus in nuclear viral factories and budding through the inner lamella of the nuclear membrane which has been modified by the insertion of herpes glycoproteins, throughout the Golgi and final release at the plasma membrane.
Latent replication : replication of circular viral episome in tandem with the host cell DNA using the host cell replication machinery.
Host-virus interaction
Adaptive immune response inhibition
Many HCMV proteins are dedicated to interfere with the host adaptive immune system. HCMV US11 reverse the translocation of human class I heavy chains from the host endoplasmic reticulum (ER) back to the cytosol which ultimately results in their degradation by the proteasome. US2 destroys two components of the MHC class II pathway, HLA-DR-alpha and DM-alpha, preventing recognition by CD4+ T cells. US3 prevents forward transport and maturation of all major histocompatibility complex (MHC) class I molecules by trapping them in the ER. HCMV US6 inhibits expression of MHC class I on the cell surface through TAP (specific transporters associated with antigen presentation) by blocking translocation of peptides from the proteasome into the ER for loading onto MHC class I.
Apoptosis modulation
Viral inhibitor of caspase-8-induced apoptosis (from gene UL36) plays a role in the inhibition of apoptosis by interacting with the pro-domain of pro-caspase-8/CASP8 and thus preventing its activation
 . Another viral protein vMIA inhibits BAX- but not BAK- mediated apoptosis.
. Another viral protein vMIA inhibits BAX- but not BAK- mediated apoptosis.
Autophagy modulation
The human cytomegalovirus protein TRS1 inhibits host autophagy process via its interaction with Beclin 1  .
.
Cell-cycle modulation
The conserved UL24 family of human alpha, beta and gamma herpesviruses induces a cell cycle arrest at G2/M transition through inactivation of the host cyclinB/cdc2 complex. HCMV encodes an UL24 homolog that should fullfill this role  .
.
Innate immune response inhibition
HCMV inhibits the cascade leading to production of interferon-beta by targeting host IRF3 protein with the viral pp65 protein  .
.
NK cells are a component of the innate immune system which play an important role in the early control of viral infections and also help to drive subsequent adaptive immunity. Several HCMV proteins are involved in the inhibition of NK-cells cytotoxicity including UL16 or UL142 that retain NKGCD2 ligands inside the cell to prevent their cell surface expression  .
.
Host splicing inhibition
HCMV UL69 modulates the host mRNA expression by exporting unspliced mRNA, thereby inducing alternative splicing  .
.
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)0 entry grouped by protein
Aotine betaherpesvirus 1 taxid:50290
Cynomolgus cytomegalovirus taxid:1919083
Human cytomegalovirus taxid:10359
Human cytomegalovirus (strain 5508) taxid:69168
| Protein | ModelArchive |
| Envelope glycoprotein L (gL) | ma-jd-viral-00341 |
