Megalocytivirus (taxid:308906)
VIRION
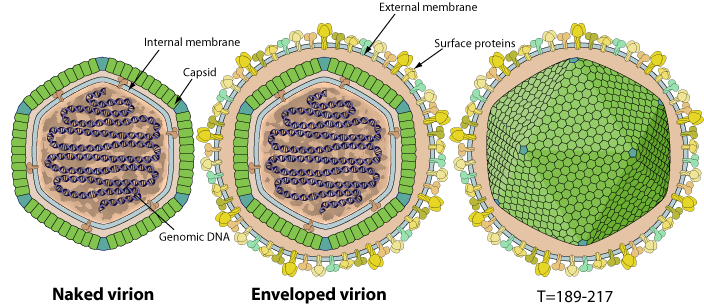
Polyhedral virions 140-200 nm in diameter. The capsid displays an icosahedral symmetry T=189-217, with an internal lipid membrane. Virions are either externally enveloped or not depending whether they budded from the cell membrane, or were arranged in paracrystaline array in the host cell cytoplasm and were released by lysis.
- Hexons: Double jelly roll-fold major capsid protein [Q77HV9]
- Pentons: Single jelly roll-fold [?]
GENOME
Linear, dsDNA genome of about 110 kb. The genome contains terminal and redundant sequences and is circularly permuted. Encodes for approximately 125 proteins.
Approximately 25% of the cytosine residues of the genome are methylated by a virus encoded DNA methyltransferase.
GENE EXPRESSION
ENZYMES
- DNA-directed DNA polymerase
- DNA-directed RNA polymerase
- Cell-type capping
- Cysteine protease (Peptidase C1A)
REPLICATION
NUCLEO-CYTOPLASMIC
- Attachment of the viral proteins to host receptors mediates endocytosis of the virus into the host cell.
- Fusion with the plasma membrane to release the DNA core into the host cytoplasm.
- Viral DNA is transported to the cell nucleus where host macromolecular synthesis is rapidly shutdown. Transcription is initiated by virally modified host RNA polymerase II.
- Parental DNA is used to produce genome and greater than genome length DNA.
- Progeny DNA is transported into cytoplasmic viral factories where large concatamers of viral DNA are formed by recombination. Transcription of very late transcripts may also take place in the cytoplasm.
- Assembly of new virions in the cytoplasm.
- Virions exit the cell by budding or cell lysis.
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)2 entries grouped by protein
1 entry
DNA polymerase (EC 2.7.7.7)
1 entry
