Cystovirus (taxid:10878)
VIRION
Enveloped, spherical virion of 85 nm in diameter. The virion has a double capsid structure: the outer capsid has a T=13 laevo icosahedral symmetry and the inner capsid has a T=2* icosahedral symmetry  .
.
Virion maturation schemes
GENOME
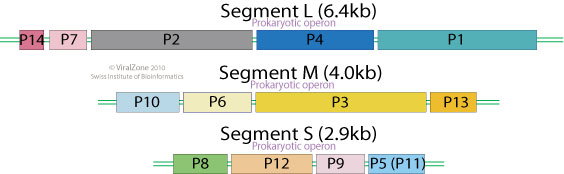
Segmented linear dsRNA genome. Contains 3 segments: Large (L), Medium (M) ans Small (S) coding for 12 proteins. Segments size range from 2.9 kb to 6.4 kb. Genome total size is 13.3 Kb.
GENE EXPRESSION
Each segments encode for a polycystronic mRNA, translated into several proteins by the procaryotic translation machinery.
ENZYMES
- RNA-dependent RNA polymerase [RdRp]
- Peptidoglycan hydrolase [gp5]
- Packaging helicase [P4]
REPLICATION
CYTOPLASMIC
- There are two different attachment processes depending on the considered species. Multimeric protein P3 adsorb to host type IV pili which retracts bringing the virion in contact with the host membrane (e.g. Phi6). In other species, the attachment apparatus is a heteromeric protein assembly that utilizes the rough lipopolysaccharide as a receptor (e.g. Phi8, Phi12 or Phi13).
- Protein P6 mediates fusion of virus membrane with host outer membrane, releasing the capsid into the periplasmic space.
- The lytic enzyme P5 on the capsid surface digests the peptidoglycan layer of the host
- The capsid enters into the cytoplasm possibly via permeabilization of the host plasma membrane. Protein P8, which may be involved in this process, dissociates from the viral capsid. The rest of the capsid is released into the host cytoplasm.
- Genomic dsRNA remains enclosed by the inner capsid, thereby "hiding" from host antiviral mechanism.
- Polycystronic mRNA are transcribed by the viral RNA-dependent RNA polymerase (RdRp) P2 inside the core particle and released into the cell cytoplasm, so that dsRNA is never exposed to the cytoplasm. Full-length plus-strand transcripts from each of the dsRNA segments are synthesized. These plus-strand transcripts are used as templates for translation.
- Neo-synthesized proteins assemble into new capsids. The packaging
NTPase P4 translocates the three ssRNA(+) genomic segments: S, M and Then L
 in the capsids where they are converted into dsRNA by the viral RdRp P2.
in the capsids where they are converted into dsRNA by the viral RdRp P2. - The capsid is matured and enveloped in the cytoplasm.
- Mature virions are released from the cell by lysis.
Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging
Juha T. Huiskonen, Felix de Haas, Doryen Bubeck, Dennis H. Bamford, Stephen D. Fuller, Sarah J. Butcher
Structure June 2006; 14: 1039-1048
Juha T. Huiskonen, Felix de Haas, Doryen Bubeck, Dennis H. Bamford, Stephen D. Fuller, Sarah J. Butcher
Structure June 2006; 14: 1039-1048
Microbiol Mol Biol Rev. 1999 Mar;63(1):149-60.
Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6
L Mindich
