dsDNA bidirectional replication
Related to: Most nuclear dsDNA viruses, and many phages.
Location: In host cell nucleus (eukaryots) or cytoplasm (prokaryots).
Replication events:
This kind of replication is used by all cellular organisms and some DNA viruses. It is the most classical way of replicating genomic nucleic acid.
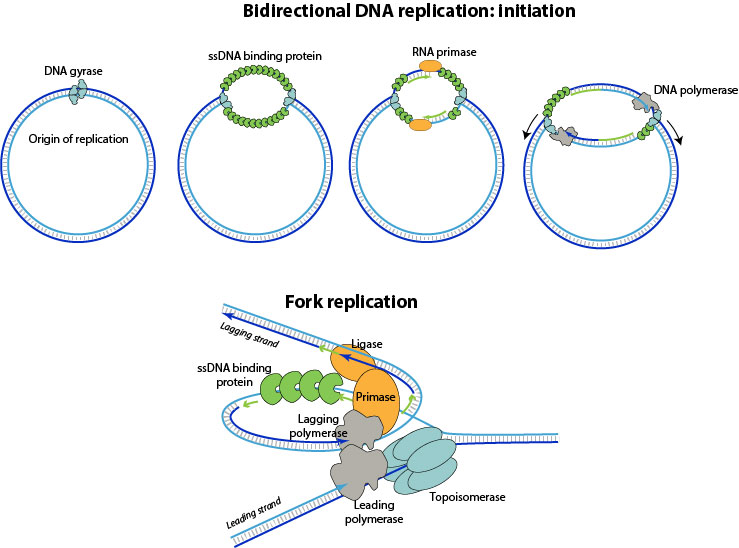
1. DNA replication begins at specific locations in the genome, called "origins". A topoisomerase unwinds the DNA double-strand at the origin of replication. ssDNA-binding proteins cover the single strand DNA created in the replication bundle.
2. A primase synthesizes short RNA primers that are then used by the DNA polymerase to prime DNA synthesis. No known enzyme is able to create a DNA primer, so all DNA synthesis begins by 3'elongation of a RNA (bidirectional replication), DNA (strand displacement, nick translation), or by protein priming (virus only).
3. The DNA polymerase and associated factors begins to elongate the leading strand at the fork. For the lagging strand Okazaki fragments !/resources/external_link.gif! are elongated after sequential RNA primer synthesis by the primase.
The lagging strand RNA primer are removed and Okazaki fragments ligated.
4. The replication forks go on until they reach the end of linear genome, or until they meet at the opposite side of a circular genome. After synthesis, topoisomerase allows separation of the two strands resulting from the replication.
| Function | Eukaryotic cell | Herpesviridae | Polyomaviridae | Papillomaviridae | Baculoviridae |
| Origin binding protein | ? | UL9 | LT | E1-E2 | IE1 |
| Topoisomerase | MCM complex | UL5 | LT | E1-E2 | p143 |
| ssDNA binding | RPA | ICP8 | host RPA | host RPA | LEF-3 |
| Primase | UL52 | host primase | host primase | LEF-1 | |
| Primase accessory factor | - | UL8 | - | - | LEF-2 |
| Polymerase, leading strand | Pol delta | UL30 | host pol delta | host pol delta | DNA pol |
| Polymerase, lagging strand | Pol epsilon | UL30 | host pol delta | ? | DNA pol |
| Processivity factor | - | UL42 | - | - | ? |
