dsDNA rolling circle replication
Related to: Most nuclear dsDNA viruses, and many phages.
Location: In host cell nucleus (eukaryots) or cytoplasm (prokaryots).
Replication events:
This kind of replication is used by many dsDNA viruses.
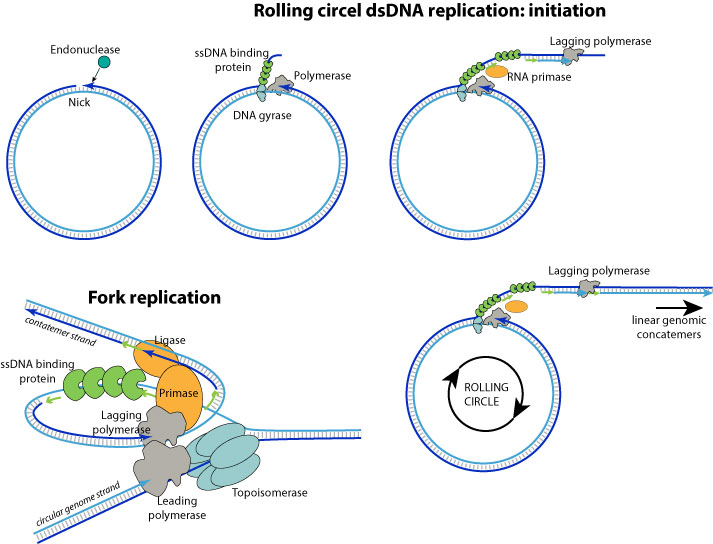
1. DNA replication begins at specific locations in the genome, called "origins". A viral endonuclease creates a nick in the origin of replication.
2. The replication machinery assembles with the DNA polymerase on the 3' extremity.
3. The DNA polymerase and associated factors begins to proceed to a strand displacement synthesis, producing a concatemer linear single stranded DNA with one genome copy per turn of replication. On the concatemer strand, Okazaki fragments are elongated after sequencial RNA primer synthesis by the primase, theus turning it into dsDNA.
The concatemer strand RNA primer are removed and okazaki fragments ligated.
4. The replication forks go on and produces a long linear concatemer which will be processed into linear genomes and encapsidated.
| Function | Herpesviridae |
| Endonuclease "Rep" | UL12? |
| Topoisomerase | UL5 |
| ssDNA binding | ICP8 |
| Primase | UL52 |
| Primase accessory factor | UL8 |
| Polymerase | UL30 |
| Processivity factor | UL42 |
