Negative-stranded RNA virus polymerase stuttering
Related to: Bornaviridae Filoviridae Paramyxoviridae Rhabdoviridae Orthomyxoviridae
Location: Host cell cytoplasm or nucleus
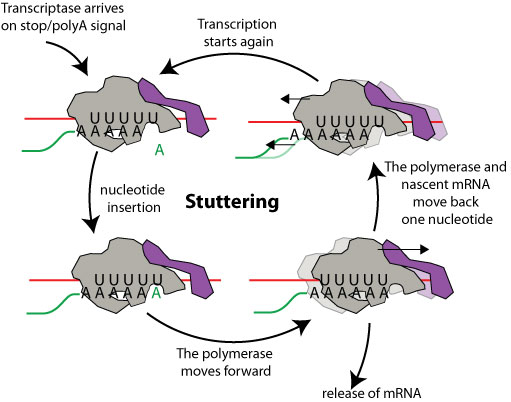
Stuttering:Many negative stranded RNA virus polyadenylate their mRNA through a polymerase stuttering mechanism during transcription. The stop signal present at the end of each gene comprises a stretch of U on which the viral polymerase acquires a stuttering behavior: after each Adenine inserted, it moves back one nucleotide along with the mRNA. It resumes transcription adding a new A, then again moves back. In this way a U on the genomic template is copied hundreds of times at the end of viral mRNA thereby producing a polyA tail. Eventually the polymerase will release the polyAdenylated mRNA and stop transcription or scan to restart on the next gene.
This process is mechanistically related to the co-transcriptional mRNA editing of paramyxoviridae and ebolaviridae.


