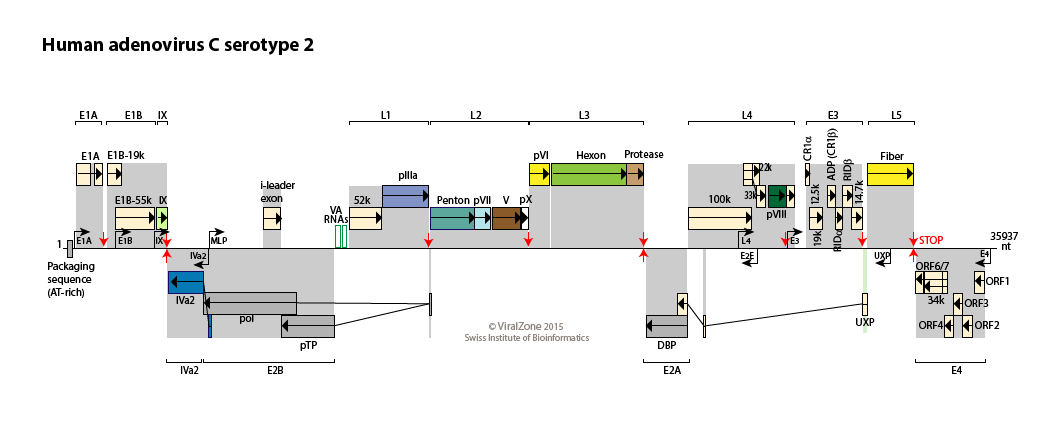
Human adenovirus type C genome




Reference sequence
Available Adenovirus sequences
Adenovirus transcription cascade
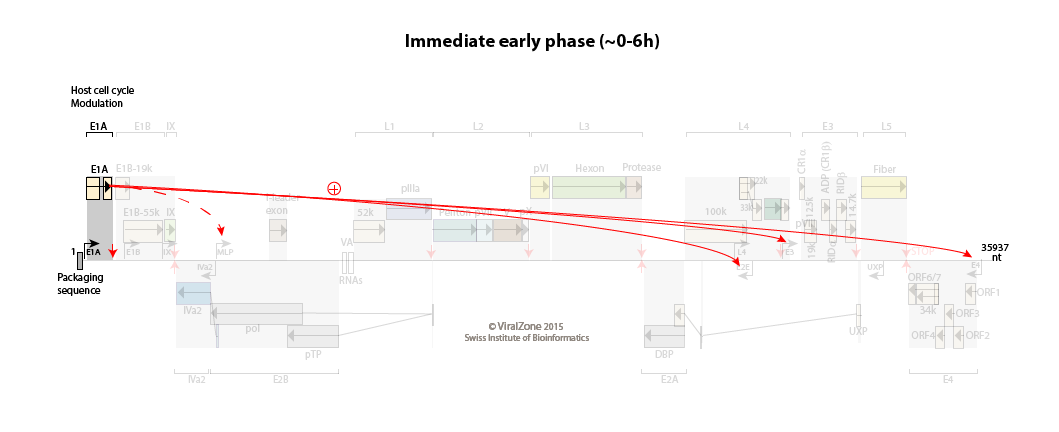
E1A is transcribed from the E1A immediate early promoter as soon as the viral genome enters the cell nucleus.
The host cell cycle is pushed into the S phase.
Immediate early to early switch: E1A activates the early promoters 
Early:
Initial expression of E1A provides transcriptional activators that, with host proteins, turn on the expression of the early promoters E1B, E2E (E2 early), E3, and E4.
The early proteins optimize the cellular milieu for viral replication, and counteract a variety of antiviral defenses.
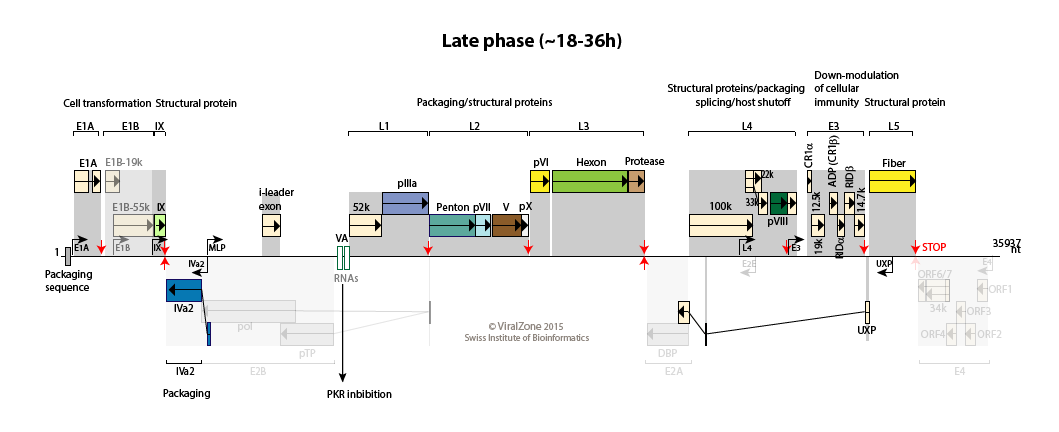
The major late promoter (MLP) is weakly active during this phase with a transcription up to L1 (primarily producing the i-leader protein and L1-52/55K proteins).
Efficient transcription from the E2E promoter results in accumulation of the E2A DNA binding protein (DBP), E2B precursor terminal protein, and DNA polymerase, which set the stage for viral DNA replication to begin.
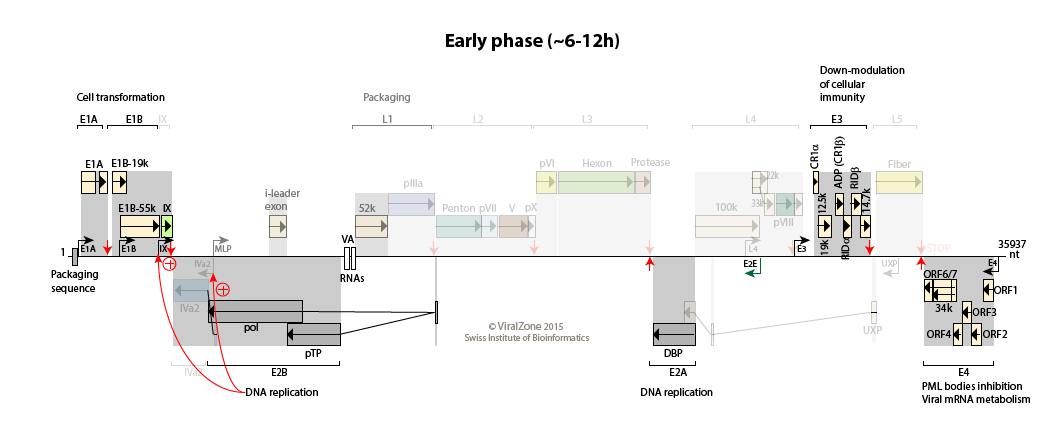
Early to intermediate switch: accumulating pool of viral genomes activates the IVa2 and IX promoters 


Activation of the transcription from pIX, IVa2, and E2 late promoters.
Transcription of the E4 region declines probably due to the repression by the DNA-binding protein.
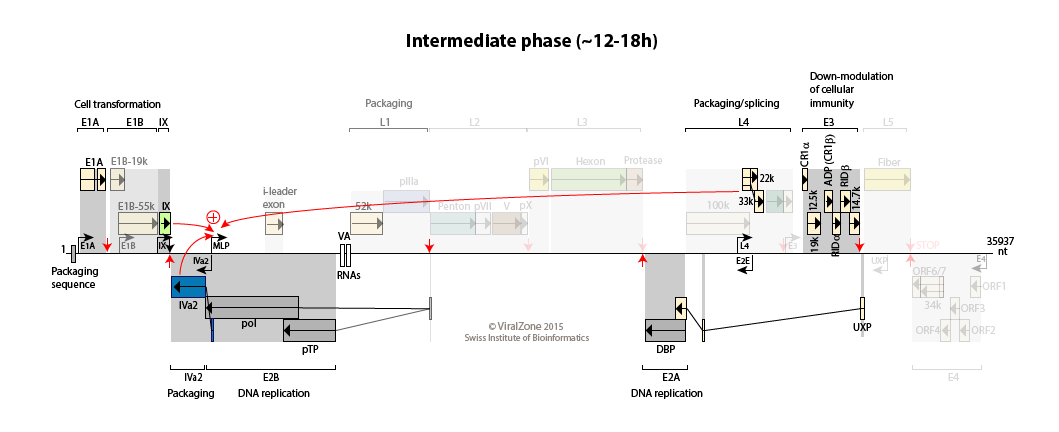
Early to late switch: E4orf4 induces late mRNA production  . pIX and IVa2 proteins upregulate the transcriptional activity of the major late promoter (MLP)
. pIX and IVa2 proteins upregulate the transcriptional activity of the major late promoter (MLP) 
 . Expression of L4-22K from L4 promoter activates maximal expression of the MLP
. Expression of L4-22K from L4 promoter activates maximal expression of the MLP 
 . L4-22K and L4-33K regulate the splicing pattern of late transcripts
. L4-22K and L4-33K regulate the splicing pattern of late transcripts  .
.
Late:
The MLP is fully activated and produces transcripts from L1 to L5 to allows synthesis of structural proteins.
100K protein induces host translation shutoff, late viral transcripts being translated efficiently through ribosomal shunting.
The adenovirus death protein (ADP) encoded by region E3 is expressed early but is greatly amplified at late stages of infection. UXP is also abundantly expressed during the late stages of infection.
Splicing events
Except for the polypeptide IX mRNA, all primary transcripts undergo one or more splicing events which give rise to about fifty distinct mRNAs during the lytic infection  .
.
Encoded proteins
| Type |
Protein name |
Function |
| Capsid shell |
Hexon protein |
T=25 icosahedral capsid, Microtubular inwards viral transport |
| Penton protein |
T=25 icosahedral capsid, Viral attachment to host host entry receptor, Clathrin-mediated endocytosis of virus by host |
| Fiber protein |
Viral attachment to host adhesion receptor |
| Hexon-linking protein IIIa |
Cement protein, Hexon stabilization
genome packaging |
| Pre-protein VI, Endosome lysis protein, Protease cofactor |
Pre-protein VI: Scaffolding of icosahedron/Nuclear capsid assembly?
Cement protein, peripentonal hexons stabilization
Endosome lysis protein: Viral penetration via permeabilization of host endosomal membrane
Protease cofactor: Virion maturation |
| Hexon-linking protein VIII |
Cement protein, peripentonal hexons stabilization |
| Hexon-interlacing protein IX |
Cement protein, hexons stabilization
Viral penetration into host nucleus |
| Packaging protein 1/IVa2 |
Present at a single vertex |
| Core proteins | Protease |
Virion maturation |
| Core-capsid bridging protein V |
Binding of the viral genome and the proteins VI and VIII thereby fixing the DNA to the capsid shell
Nuclear capsid assembly? |
| Histone-like nucleoprotein VII |
Histone-like nucleoprotein, DNA binding/condensation |
| Core protein X | Nucleoprotein, DNA binding/condensation |
| Replication proteins |
Terminal protein |
Strand displacement replication (DNA synthesis priming) |
DBP | Strand displacement replication (DNA unwinding) |
Polymerase | Strand displacement replication (DNA synthesis) |
33K protein | Viral alternative splicing |
| Packaging |
Packaging protein 1/IVa2 |
Packaging (ATPase motor) |
| Packaging protein 2/22K protein | Packaging |
| Packaging protein 3/52K | Packaging |
33K protein | Packaging (Terminase) |
Host-modulation proteins | Shutoff protein 100K | Host translation shutoff., Ribosomal shunting, Scaffolding (hexons trimerization) |
| Early E1A 32 kDa protein | G1/S host cell cycle checkpoint dysregulation by virus, cell transformation, Inhibition of host transcription initiation, Inhibition of STAT1 activity |
| E1B 19 kDa protein |
Inhibition of host apoptosis by viral BCL2-like protein |
| E1B 55 kDa protein (TP53_E4-Orf3_E4-Orf6_PML) | Inhibition of p53 and E1A-induced apoptosis during the oncogenic transformation induced by the virus |
| E3 12.5k protein | Unknown |
| E3 CR1-α protein | apoptosis modulation (cell survival) by TRAIL downregulation and/or ER regulation |
| E3 18.5 kDa glycoprotein (gp19K) | MHC escape: Inhibition of host tapasin by virus |
| Adenovirus death protein / CR1-β | cell lysis |
| E3 RIDα protein | Apoptosis modulation (cell survival) by down-regulation of the TNF-family death receptors |
| E3 RIDβ protein | Apoptosis modulation (cell survival) by down-regulation of the TNF-family death receptors |
| E3 14.7k protein | Inhibition of host NF-kappa-B? |
| E4 ORF1 protein | ?tight junction modulation? PDZ protein binding? |
| E4 ORF2 protein | Unknown |
| E4 ORF3 protein | PML bodies inhibition |
| E4 ORF4 protein | ?alternative splicing? |
| E4 ORF 34k protein | Nuclear innate immunity inhibition,PML bodies inhibition? |
| E4 ORF6/7 control protein | switch to intermediate phase, G1/S host cell cycle checkpoint dysregulation by virus |
| I-leader protein | Unknown |
| U exon | unknown |
Variation of the E3 region among human AdV species
From: Robinson et al. "Molecular evolution of
human species D adenoviruses." Infect Genet Evol. 11:1208-17 (2011). 
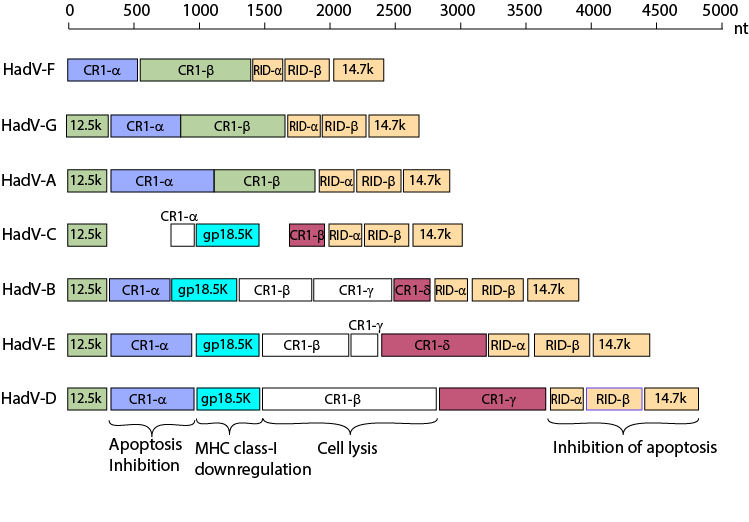
Molecular evolution of human species D adenoviruses
Christopher M. Robinson, Donald Seto, Morris S. Jones, David W. Dyer, James Chodosh
Infect. Genet. Evol. August 2011; 11: 1208-1217
A new look at adenovirus splicing
Hongxing Zhao, Maoshan Chen, Ulf Pettersson
Virology May 2014; 456-457: 329-341
Promoter occlusion prevents transcription of adenovirus polypeptide IX mRNA until after DNA replication
L. D. Vales, J. E. Darnell
Genes Dev. January 1989; 3: 49-59
Adenovirus 2 peptide IX gene is expressed only on replicated DNA molecules
T. Matsui, M. Murayama, T. Mita
Mol. Cell. Biol. December 1986; 6: 4149-4154
Regulation of early adenovirus gene expression
J. R. Nevins
Microbiol. Rev. December 1987; 51: 419-430
Adenovirus L4-22K stimulates major late transcription by a mechanism requiring the intragenic late-specific transcription factor-binding site
Ellenor Backstrom, Kerstin B. Kaufmann, Xin Lan, Goran Akusjarvi
Virus Res. August 2010; 151: 220-228
Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter
P. Lutz, C. Kedinger
J. Virol. March 1996; 70: 1396-1405
Viral DNA synthesis-dependent titration of a cellular repressor activates transcription of the human adenovirus type 2 IVa2 gene
C. Iftode, S. J. Flint
Proc. Natl. Acad. Sci. U.S.A. December 21, 2004; 101: 17831-17836
The product of the adenovirus intermediate gene IX is a transcriptional activator
P. Lutz, M. Rosa-Calatrava, C. Kedinger
J. Virol. July 1997; 71: 5102-5109
Regulation of human adenovirus alternative RNA splicing by the adenoviral L4-33K and L4-22K proteins
Roberta Biasiotto, G?ran Akusj?rvi
Int J Mol Sci 2015; 16: 2893-2912
The adenovirus L4-33K protein regulates both late gene expression patterns and viral DNA packaging
Kai Wu, Diana Guimet, Patrick Hearing
J. Virol. June 2013; 87: 6739-6747
Adenovirus E4orf4 induces HPV-16 late L1 mRNA production
Monika Somberg, Margaret Rush, Joanna Fay, Fergus Ryan, Helen Lambkin, G?ran Akusj?rvi, Stefan Schwartz
Virology January 20, 2009; 383: 279-290




 . pIX and IVa2 proteins upregulate the transcriptional activity of the major late promoter (MLP)
. pIX and IVa2 proteins upregulate the transcriptional activity of the major late promoter (MLP) 
 . Expression of L4-22K from L4 promoter activates maximal expression of the MLP
. Expression of L4-22K from L4 promoter activates maximal expression of the MLP 
 . L4-22K and L4-33K regulate the splicing pattern of late transcripts
. L4-22K and L4-33K regulate the splicing pattern of late transcripts  .
.
 .
.