RNA suppression of termination (kw:KW-1159)
Suppression of termination (also referred to as stop codon readthrough) is an alternate mechanism of translation to produce a protein with an extended C-terminus. It is used as a regulatory strategy by a large number of plant, animal and prokaryot viruses (see table below).
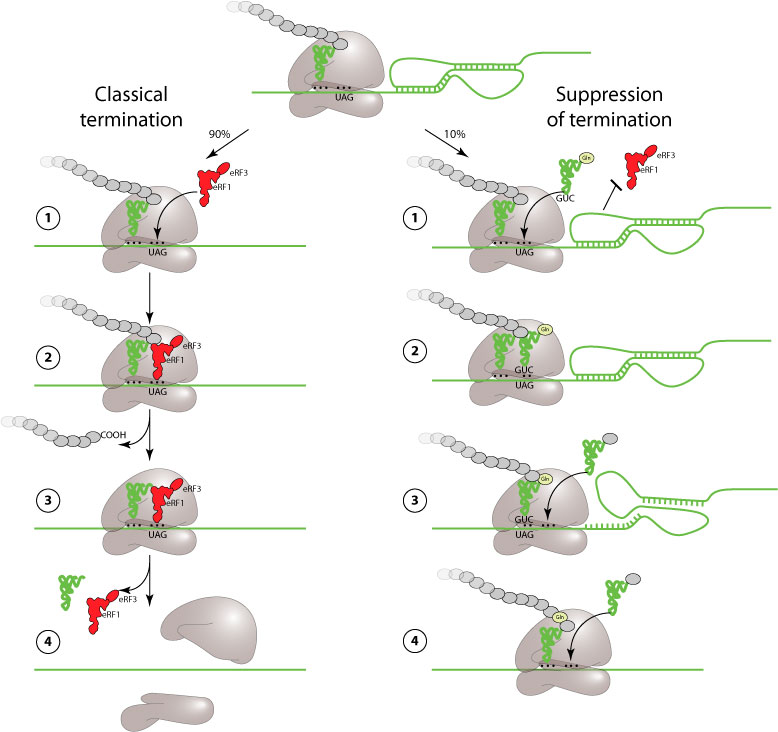
UAG amber and UGA opal codons are normally recognized by a release factor (eRF) instead of a tRNA, thereby stopping translation. Readthrough of these stop codons occurs when a tRNA partially recognizes UGA or UAG and allows translation to proceed further. This readthrough is induced either by a pseudoknot (MMTV) or by few nucleotides sequence after the stop. The readthrough signals of prokaryote, plant and mammal viruses are different and have different mechanisms which are still poorly understood. The efficiency of suppression of termination is globally poor: 5 to 30%.
The picture below describes MMTV suppression of termination.
| Family | Genus/TD> | Virus | RefSeq | STOP codon | Insertion | Efficiency | Position | Reference |
| Fiersviridae | qubevirus | Enterobacteria phage Qbeta | NC_001890 | UGA (opal) | Trp | 5% | 1742 |  |
| Carmotetraviridae | Alphacarmotetravirus | Providence virus | NC_014126 | UAG (amber) | - | - | 2104 |  |
| Luteoviridae | Enamovirus | Pea enation mosaic virus 1 | NC_003629 | UGA (opal) | - | - | 4566 |  |
| Luteovirus | Barley yellow dwarf virus | NC_004750 | UAG (amber) | - | - | 3458 |  |
|
| Polerovirus | Potato leafroll virus | NC_001747 | UAG (amber) | - | - | 4317 |  |
|
| Reoviridae | Coltivirus | Colorado tick fever virus | NC_004180 | UGA (opal) | - | - | 1052 |  |
| Retroviridae | Gammaretrovirus | Moloney murine leukemia virus | NC_001501 | UAG (amber) | Gln | 4-10% | 1974 |  |
| Epsilonretrovirus | Walleye dermal sarcoma virus | NC_001867 | UAG (amber) | - | - | 2545 | ||
| Togaviridae | Alphavirus | Sindbis virus | NC_001547 | UGA (opal) | - | 10% | 7598 |  |
| Tombusviridae | Aureusvirus | Pothos latent virus | NC_000939 | UAG (amber) | - | - | 791 |  |
| Avenavirus | Oat chlorotic stunt virus | NC_003633 | UAG (amber) | - | - | 684 |  |
|
| Carmovirus | Carnation mottle virus | NC_001265 | UAG (amber) | - | - | 805 | ||
| Machlomovirus | Maize chlorotic mottle virus | NC_003627 | UAG (amber) | - | - | 1451 | ||
| UGA (opal) | - | - | 3199 | |||||
| Alphanecrovirus | Tobbaco necrosis virus A | NC_001777 | UAG (amber) | - | - | 666 | ||
| Panicovirus | Panicium mosaic virus | NC_002598 | UAG (amber) | - | - | 1304 | ||
| Tombusvirus | Tomato bushy stunt virus | NC_001554 | UAG (amber) | - | - | 1054 |  |
|
| Virgaviridae | Furovirus | Soil-borne wheat mosaic virus | NC_001367 | UGA (opal) | - | - | 4032 | |
| NC_002042 | UGA (opal) | - | - | 862 |  |
|||
| Pecluvirus | Peanut clump virus | NC_003672 | UGA (opal) | - | - | 3567 | ||
| Pomovirus | Potato mop-top virus | NC_003723 | UGA (opal) | - | - | 4024 | ||
| NC_003724 | UAG (amber) | - | - | 842 |  |
|||
| Tobamovirus | Tobacco mosaic virus | NC_002041 | UGA (opal) | Tyr, Gln, Leu | 31% | 3417 |  |
|
| Tobravirus | Tobacco rattle virus | NC_003805 | UGA (opal) | Trp | 5-25% | 3764 |  |
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)78 entries grouped by protein
2 entries
Minor capsid protein A1
4 entries
Readthrough protein P3-RTD (P3-P5 readthrough protein) (P74) (Readthrough protein) (RT protein)
1 entry
Protein p31
13 entries
Gag-Pol polyprotein
25 entries
Polyprotein P1234 (P1234) (Non-structural polyprotein)
32 entries
Replicase large subunit (EC 2.1.1.-) (EC 2.7.7.-) (EC 2.7.7.48) (EC 3.6.4.13) (183 kDa protein) (RNA-directed RNA polymerase)
1 entry
