Curtovirus (taxid:10813)
VIRION
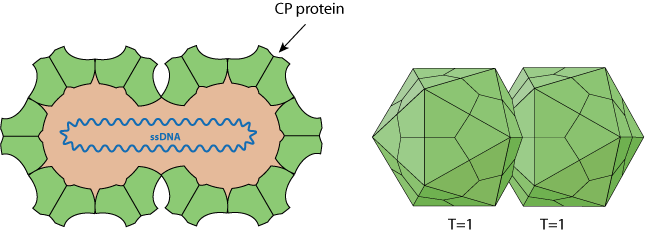
Non-enveloped, about 38 nm in length and 22 nm in diameter (for MSV), twinned (geminate) incomplete T=1 icosahedral symmetry capsid that contains 22 pentameric capsomers made of 110 capsid proteins (CP). Each geminate particle contains only a single circular ssDNA.
GENOME
Monopartite, circular, ssDNA genome (+) genome of about 3.0 kb. 3' terminus has no poly(A) tract. There are coding regions in both the virion (positive) and complementary (negative) sense strands.
The genome is replicated through double-stranded intermediates. The replication (Rep) protein initiates and terminates rolling circle replication, the host DNA polymerase being used for DNA replication itself. There is a potential stem-loop structure in the intergenic region that includes a conserved nona-nucleotide sequence (TAATATTAC) where ssDNA synthesis is initiated.
GENE EXPRESSION
Transcription is bidirectional from the intergenic region. 7 proteins are produced. On the v-sense: Movement protein (MP), CP and V2 protein. On the C-sense: Rep, C2, Replication enhancer (REn) and C4.
Proteins are expressed from subgenomic RNAs (sgRNAs).
ENZYMES
REPLICATION
NUCLEAR
- Virus penetrates into the host cell.
- Uncoating, the viral ssDNA genome penetrates into the nucleus.
- The ssDNA is converted into dsDNA with the participation of cellular factors.
- bidirectional dsDNA transcription from the IR promoter produces viral mRNAs and translation of viral proteins.
- Replication is initiated by cleavage of the(+)strand by REP, and occurs by rolling circle producing ssDNA genomes.
- These newly synthesized ssDNA can either
a) be converted to dsDNA and serve as a template for transcription/replication
b) be encapsidated by capsid protein and form virions released by cell lysis
c) be transported outside the nucleus, to a neighboring cell through plasmodesmata (cell-cell movement) with the help of viral movement proteins.
Host-virus interaction
Cell-cycle modulation
Curtovirus protein Rep is probably responsible for inhibiting host retinoblastoma protein and inducing transition from the G1 to S phase in preparation for virus replication since the virus targets differentiated non-dividing cells.
 .
.
Suppression of host silencing-mediated antiviral defense
Curtovirus viral proteins are able to suppress host cell RNA silencing
either by inhibiting host-mediated methylation of viral DNA
 or by inhibiting post-translational gene silencing (PTGS)
or by inhibiting post-translational gene silencing (PTGS)
 .
.
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)7 entries grouped by protein
1 entry
Protein C4 (Protein L4)
1 entry
Protein C2 (Protein L2)
1 entry
Capsid protein (Coat protein) (CP)
1 entry
Movement protein (MP)
1 entry
Replication enhancer (REn) (16.1 kDa protein) (Protein C3) (Protein L3)
1 entry
Replication-associated protein (Rep) (EC 2.7.7.-) (EC 3.1.21.-) (40.8 kDa protein) (Protein C1)
1 entry
