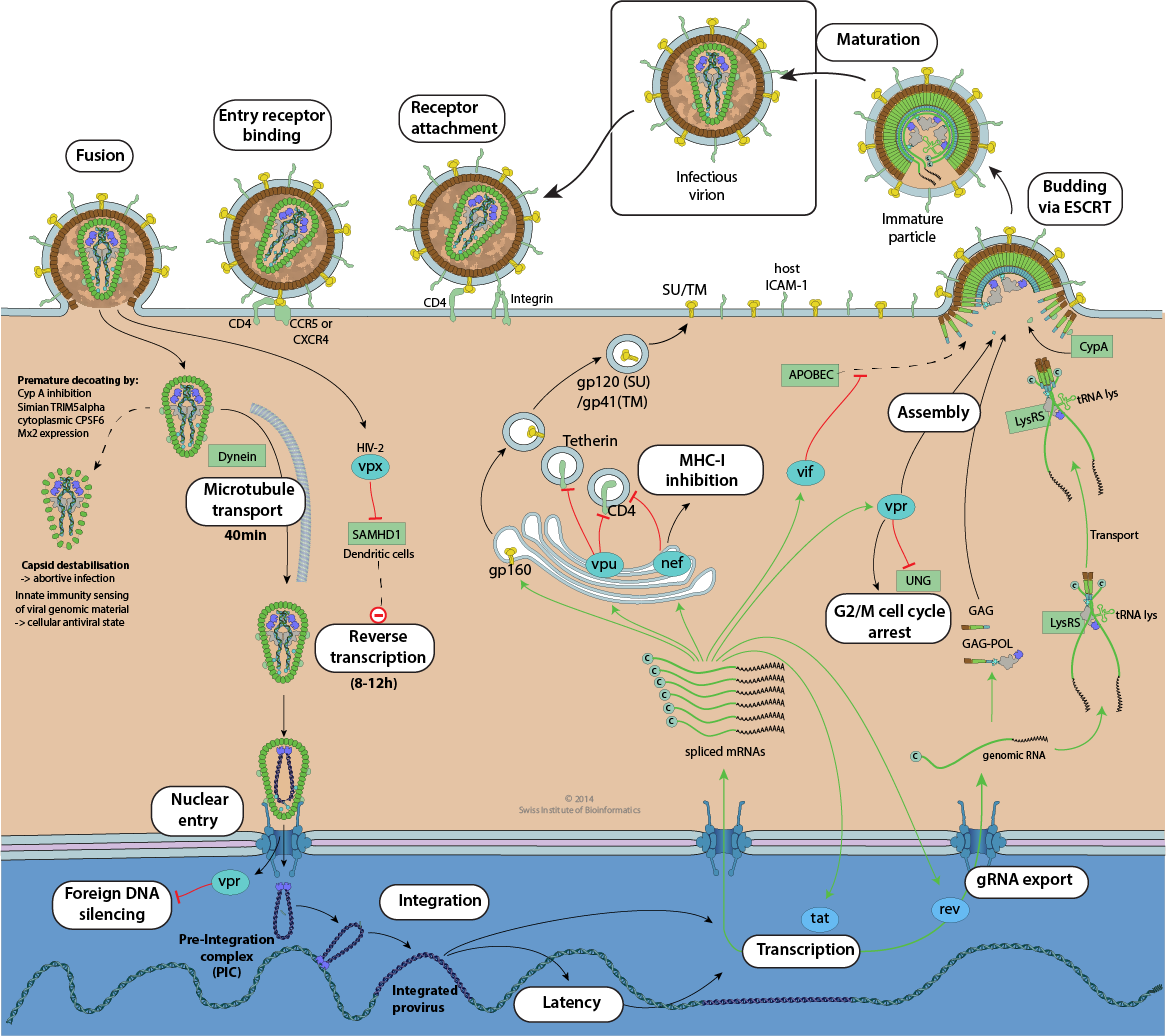
HIV replication cycle
The average lifespan of virus-producing cells is short ~2 days.

X. Wei, S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn
Nature January 12, 1995; 373: 117?122
Cycle timing:
- Reverse transcription takes about 8-12h - DNA starts to be detectable in the nucleus 4h after infection.
- DNA starts to be detectable in the nucleus 4h after infection.
- Later, the assembly of the gag-shell takes about 10 min  -Budding occurs 15min after
-Budding occurs 15min after
Attachment
The gp120 binds to the attachment receptor CD4. This binding is reversible and important for HIV entry, although not mandatory. Gp120 then binds to the entry receptor CCR5, which is a signaling receptor. It is unclear whether the virus fuses at the cell surface or is endocytosed by clathrin mecanism  .
As AIDS disease progresses, viral variant that utilize the host entry receptor CXCR4 appear, coinciding with the immunologically deficient phase of infection
.
As AIDS disease progresses, viral variant that utilize the host entry receptor CXCR4 appear, coinciding with the immunologically deficient phase of infection  Viral attachment to host cell, Fusion of virus membrane with host cell membrane
Viral attachment to host cell, Fusion of virus membrane with host cell membrane
Chris Verhofstede, Monique Nijhuis, Linos Vandekerckhove
Curr Opin HIV AIDS. 2012 Sep;7(5):432-9
Release of viral components into the cell cytoplasm.
After virus-host membrane fusion, the viral capsid is released into the cytoplasm. In certain cells, such as dendritic cells, Vpx (HIV-2 only) is thought to be transported into the nucleus, where it facilitates reverse transcription/ PIC entry by counteracting the specific host defense mechanisms of these cells.
Reverse transcription
Linear reverse transcription is initiated in the mature virion, but usually occurs after the capsid is released into the cytoplasm.
All HIV capsids are diploid, and comprise two copies of genomic RNA. Does diploidy play a role in reverse transcription?
Several studies have addressed this question: First strand transfer could occur intermolecularly  , but it could also occur with a single molecule of genomic RNA
, but it could also occur with a single molecule of genomic RNA 
 . In other words, the mechanism of reverse transcription does not explain the observed diploidy of the capsid.
. In other words, the mechanism of reverse transcription does not explain the observed diploidy of the capsid.
A. T. Panganiban, D. Fiore
Science August 26, 1988; 241: 1064?1069
J. L. van Wamel, B. Berkhout
Virology May 10, 1998; 244: 245?251
J. S. Jones, R. W. Allan, H. M. Temin
J. Virol. January 1994; 68: 207?216
Transport
The viral capsid interacts with microtubules and is actively transported near the nucleus. This would occur simultaneously with reverse transcription. This transport takes 40 min ex vivo 
 .
Microtubular inward viral transport
.
Microtubular inward viral transport
Rapha?l Gaudin, Bruna Cunha de Alencar, Nathalie Arhel, Philippe Benaroch
Trends Cell Biol. December 2013; 23: 652?662
Uncoating and nuclear entry
Uncoating is the process by which the viral cDNA genome is released from the capsid. It is a multistep process closely associated with reverse transcription  . Early uncoating can occur under certain circumstances and leads to termination of infection: CypA depletion, simian Trim5alpha binding
. Early uncoating can occur under certain circumstances and leads to termination of infection: CypA depletion, simian Trim5alpha binding , Mx2 antiviral protein expression
, Mx2 antiviral protein expression or cytoplasmic CPSF6
or cytoplasmic CPSF6 . Early uncoating may induce an antiviral response of the cytoplasmic dsDNA sensor.
. Early uncoating may induce an antiviral response of the cytoplasmic dsDNA sensor.
Entry of the pre-integration complex (cDNA, integrase) into the nucleus occurs through various bindings to the nuclear pore and is dependent on the presence of vpr.
Viral penetration into host nucleus
Zandrea Ambrose, Christopher Aiken
Virology April 2014; 454-455: 371?379
Emi E. Nakayama, Tatsuo Shioda
Rev. Med. Virol. March 2010; 20: 77?92
KyeongEun Lee, Zandrea Ambrose, Thomas D. Martin, Ilker Oztop, Alok Mulky, John G. Julias, Nick Vandegraaff, Joerg G. Baumann, Rui Wang, Wendy Yuen, Taichiro Takemura, Kenneth Shelton, Ichiro Taniuchi, Yuan Li, Joseph Sodroski, Dan R. Littman, John M. Coffin, Stephen H. Hughes, Derya Unutmaz, Alan Engelman, Vineet N. KewalRamani
Cell Host Microbe March 18, 2010; 7: 221?233
Caroline Goujon, Olivier Moncorg?, H?l?ne Bauby, Tomas Doyle, Christopher C. Ward, Torsten Schaller, St?phane Hu?, Wendy S. Barclay, Reiner Schulz, Michael H. Malim
Nature October 24, 2013; 502: 559?562
Integration
The viral cDNA is covalently integrated into the host genome by integrase, preferentially at transcriptionally active sites. The integrated viral genome is referred to as a provirus. Viral genome integration
Latency/ replication decision
The provirus can be transcribed to the point where it reaches tat expression and can enter the replicative phase.
Alternatively, it can be methylated by host factors, and this transcriptional silencing induces viral latency  .
Viral latency
.
Viral latency
Debbie S. Ruelas, Warner C. Greene
Cell October 24, 2013; 155: 519?529
Latency/ reactivation
Cellular stress or activation can lead to nuclear signals that abrogate transcriptional silencing of the provirus, initiating the viral replication cycle  .
.
Kotaro Shirakawa, Leonard Chavez, Shweta Hakre, Vincenzo Calvanese, Eric Verdin
Trends Microbiol. June 2013; 21: 277?285
Transcription
Provirus transcription is under the control of tat and many host transcription factors that interact with the U3 promoter. mRNA transcripts can be alternatively spliced into six mRNAs. dsDNA templated transcription
Genomic RNA exit from the nucleus
Unspliced genomic RNA (gRNA) cannot be exported to the cytoplasm under normal circumstances, because unspliced RNA is prevented from doing so in eukaryotes. The viral protein rev specifically unlocks gRNA export to the cytoplasm. The gRNA serves both as a template for GAG and POL translation and as genomic material for budding into new virions. Genome export through nuclear pore
Virion assembly and budding
The genomic packaging signal (Psi) is only encoded in genomic RNA (Psi on wikipedia). In monomeric gRNA, the Psi element is inactive because it is sequestered by intramolecular base pairing. After gRNA dimerization, cross-hybridization of the RNA exposes the Psi element, which allows specific binding of the gRNA dimer to the nucleoprotein (NC) domain, and initiates assembly .
Subsequently, several GAG proteins interact with a gRNA-NC complex. The latter is transported by actin or microtubules to the plasma membrane, where final virion assembly and budding occurs.
.
Subsequently, several GAG proteins interact with a gRNA-NC complex. The latter is transported by actin or microtubules to the plasma membrane, where final virion assembly and budding occurs.
 Viral budding via the host ESCRT complexes
Viral budding via the host ESCRT complexes
Maturation of the virion
Upon release of the viral particle, the protease is activated and cleaves the polyproteins GAG and GAG - POL into individual chains that self-assemble in the virion to form a conical capsid. In some virions the capsid may self-assemble into a cylinder, in these the viral genome appears to be from the formation of the capsid  .
After maturation, all elements are in place to allow reverse transcription. NC would play a role in orchestrating the timing of maturation-reverse transcription, as some NC mutants trigger these events before budding and turn HIV into a DNA virus
.
After maturation, all elements are in place to allow reverse transcription. NC would play a role in orchestrating the timing of maturation-reverse transcription, as some NC mutants trigger these events before budding and turn HIV into a DNA virus  .
Virion maturation
.
Virion maturation
The HIV resource is supported by the Swiss South Africa Joint Research Programme.
Cora L. Woodward, Sarah N. Cheng, Grant J. Jensen
J. Virol. November 12, 2014;
Laurent Houzet, Zakia Morichaud, Ludovic Didierlaurent, Delphine Muriaux, Jean-Luc Darlix, Maryl?ne Mougel
Nucleic Acids Res. April 2008; 36: 2311?2319
M?lanie R. Tardif, Michel J. Tremblay
J. Virol. November 2003; 77: 12299?12309
E. K. Franke, H. E. Yuan, J. Luban
Nature November 24, 1994; 372: 359?362
Takeo Dochi, Takashi Nakano, Mutsumi Inoue, Nobutoki Takamune, Shozo Shoji, Kouichi Sano, Shogo Misumi
J. Gen. Virol. May 2014; 95: 1156?1166
B. M?ller, U. Tessmer, U. Schubert, H. G. Kr?usslich
J. Virol. October 2000; 74: 9727?9731
Lars-Anders Carlson, John A. G. Briggs, B?rbel Glass, James D. Riches, Martha N. Simon, Marc C. Johnson, Barbara M?ller, Kay Gr?newald, Hans-Georg Kr?usslich
Cell Host Microbe December 11, 2008; 4: 592?599
Marc Permanyer, Ester Ballana, Jos? A. Est?
Trends Microbiol. December 2010; 18: 543?551
Sergey Ivanchenko, William J. Godinez, Marko Lampe, Hans-Georg Kr?usslich, Roland Eils, Karl Rohr, Christoph Br?uchle, Barbara M?ller, Don C. Lamb
PLoS Pathog. November 2009; 5: e1000652
Nolwenn Jouvenet, Paul D. Bieniasz, Sanford M. Simon
Nature July 10, 2008; 454: 236?240
Victoria D?Souza, Michael F. Summers
Nat. Rev. Microbiol. August 2005; 3: 643?655
Ping Zhu, Elena Chertova, Julian Bess, Jeffrey D. Lifson, Larry O. Arthur, Jun Liu, Kenneth A. Taylor, Kenneth H. Roux
Proc. Natl. Acad. Sci. U.S.A. December 23, 2003; 100: 15812?15817
Thomas Wilk, Stephen D Fuller
Current Opinion in Structural Biology April 1999; 9: 231?243
John A. G. Briggs, Martha N. Simon, Ingolf Gross, Hans-Georg Kr?usslich, Stephen D. Fuller, Volker M. Vogt, Marc C. Johnson
Nat. Struct. Mol. Biol. July 2004; 11: 672?675
Zandrea Ambrose, Christopher Aiken
Virology April 2014; 454-455: 371?379
S. P. Singh, D. Lai, M. Cartas, D. Serio, R. Murali, V. S. Kalyanaraman, A. Srinivasan
Virology March 15, 2000; 268: 364?371
Sook-Kyung Lee, Marc Potempa, Ronald Swanstrom
J. Biol. Chem. November 30, 2012; 287: 40867?40874
Sergey Iordanskiy, Reem Berro, Maria Altieri, Fatah Kashanchi, Michael Bukrinsky
Retrovirology 2006; 3: 4
David McDonald, Marie A. Vodicka, Ginger Lucero, Tatyana M. Svitkina, Gary G. Borisy, Michael Emerman, Thomas J. Hope
J. Cell Biol. November 11, 2002; 159: 441?452
Salah Edin El Meshri, Denis Dujardin, Julien Godet, Ludovic Richert, Christian Boudier, Jean Luc Darlix, Pascal Didier, Yves M?ly, Hugues de Rocquigny
J. Mol. Biol. March 27, 2015; 427: 1480?1494
