Restriction-modification system evasion by virus (kw:KW-1258)
The restriction-modification (RM) system is a defense mechanism present in over 90% of sequenced bacterial and archeal genomes. Its consists of a modification enzyme that methylates a specific DNA sequence in a genome and a restriction endonuclease that cleaves DNA lacking this methylation  .<div id="PMID: 11782494" class="hidden">2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self
.<div id="PMID: 11782494" class="hidden">2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self
Noreen E Murray
Microbiology (Reading, Engl.) January 2002; 148: 3?20
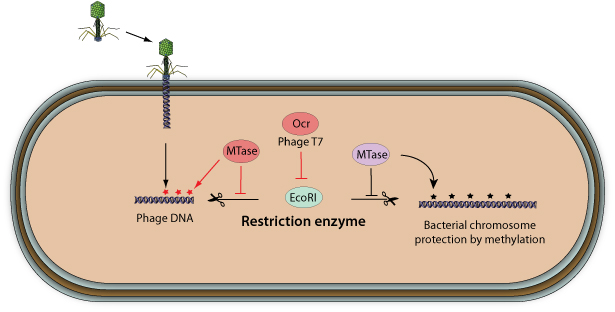
Bacterial viruses have evolved different strategies to evade the restriction-modification system 
 . Some viruses encode their own methyltransferase in order to protect their genome from host restriction enzymes
. Some viruses encode their own methyltransferase in order to protect their genome from host restriction enzymes  . Instead, bacteriophage T7 encodes the OCR protein that blocks the active site of several restriction enzymes by mimicking the phosphate backbone of B-form DNA. Other bacterial viruses use unusual bases on their genome to avoid restriction. Bacteriophages SPO1, SP82, and 2C replace thymidine with 5-hydroxymethyluracil while phages PBS1 and PBS2 thymine is completely changed to uracil.
. Instead, bacteriophage T7 encodes the OCR protein that blocks the active site of several restriction enzymes by mimicking the phosphate backbone of B-form DNA. Other bacterial viruses use unusual bases on their genome to avoid restriction. Bacteriophages SPO1, SP82, and 2C replace thymidine with 5-hydroxymethyluracil while phages PBS1 and PBS2 thymine is completely changed to uracil.
| Family | Genus | Virus | Strategy of RM evasion | Viral protein | Ref. |
| Siphoviridae | Tunalikevirus | Bacteriophage T1 | Viral methyltransferase | Dmt methyltransferase |   |
| Podoviridae | T7likevirus | Bacteriophage T3 | Viral SAMase | S-adenosyl-L-methionine hydrolase |  |
| Myoviridae | T4likevirus | Bacteriophage T4 | Viral methyltransferase | Dam methyltransferase |  |
| Myoviridae | T4likevirus | Bacteriophage T2 | Viral methyltransferase | Dam methyltransferase |  |
| Podoviridae | T7likevirus | Bacteriophage T7 | Inhibition of type-I RM enzymes | OCR protein |  |
| Myoviridae | Punalikevirus | Bacteriophage P1 | Internal capsid protein | DarA, DarB |  |
| Podoviridae | P22likevirus | Bacteriophage P22 | Modification of host methylase activity | Ral |  |
| Siphoviridae | Lambdalikevirus | Bacteriophage λ reverse | Modification of host methylase activity | Lar |  |
| Siphoviridae | Lambdalikevirus | Bacteriophage λ | Modification of host methylase activity | Ral |  |
| Myoviridae | Mulikevirus | Bacteriophage Mu | Adenine acetyltransferase | Mom |   |
Michael R Hall, Edward McGillicuddy, Lewis J Kaplan
Surg Infect (Larchmt) January 29, 2014;
Simon J Labrie, Julie E Samson, Sylvain Moineau
Nat. Rev. Microbiol. May 2010; 8: 317-327
Julie E Samson, Alfonso H Magadan, Mourad Sabri, Sylvain Moineau
Nat. Rev. Microbiol. October 2013; 11: 675-687
B Auer, M Schweiger
J. Virol. February 1984; 49: 588-590
F W Studier, N R Movva
J. Virol. July 1976; 19: 136-145
V G Kossykh, S L Schlagman, S Hattman
J. Biol. Chem. June 16, 1995; 270: 14389-14393
C Atanasiu, T-J Su, S S Sturrock, D T F Dryden
Nucleic Acids Res. September 15, 2002; 30: 3936-3944
S Iida, M B Streiff, T A Bickle, W Arber
Virology March 1987; 157: 156-166
A V Semerjian, D C Malloy, A R Poteete
J. Mol. Biol. May 5, 1989; 207: 1-13
G King, N E Murray
Gene May 19, 1995; 157:225
I TAKAHASHI, J MARMUR
Nature February 23, 1963; 197: 794-795
Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA
Proc Natl Acad Sci U S A. 1983 Dec;80(24):7400-4
James Murphy, Jennifer Mahony, Stuart Ainsworth, Arjen Nauta, Douwe van Sinderen
Appl Environ Microbiol December 2013; 79: 7547-7555
E. F. Wagner, B. Auer, M. Schweiger
J. Virol. March 1979; 29: 1229-1231
V. G. Kossykh, S. L. Schlagman, S. Hattman
J. Bacteriol. May 1997; 179: 3239-3243
Naofumi Handa, Ichizo Kobayashi
J. Bacteriol. November 2005; 187: 7362-7373
W. A. Loenen, N. E. Murray
J. Mol. Biol. July 5, 1986; 190: 11-22
A. Toussaint
Virology March 1976; 70: 17-27
F. W. Studier
J. Mol. Biol. May 15, 1975; 94: 283-295
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)45 entries grouped by protein
1 entry
Anti-restriction endonuclease (Anti-rgl nuclease)
1 entry
5-NmdU N-acetyltransferase
3 entries
DNA N-6-adenine-methyltransferase (DAM) (EC 2.1.1.72) (EcoP1I DNA MTase) (M.EcoP1I) (Type II methyltransferase M.EphP1ORF2P) (M.EphP1ORF2P)
1 entry
Defense against restriction protein A (DarA)
1 entry
Defense against restriction protein B (EC 3.6.4.-) (DarB)
3 entries
Deoxycytidylate 5-hydroxymethyltransferase (Deoxycytidylate hydroxymethylase) (EC 2.1.2.8) (HMase) (dCMP hydroxymethylase) (dCMP HMase)
2 entries
DNA adenine methylase (EC 2.1.1.72) (DNA-(N(6)-adenine)-methyltransferase) (Deoxyadenosyl-methyltransferase) (Orphan methyltransferase M.EcoT2Dam) (M.EcoT2Dam)
2 entries
Deoxynucleotide monophosphate kinase (DNK) (dNMP kinase) (EC 2.7.4.-) (Gp1)
1 entry
DNA-directed DNA polymerase (EC 2.7.7.7)
3 entries
Deoxyuridylate hydroxymethyltransferase (Deoxyuridylate hydroxymethylase) (EC 2.1.2.-) (Gp29) (dUMP hydroxymethylase) (dUMP-HMase)
1 entry
Flavin-dependent lyase (gp47)
2 entries
Glycinyltransferase (Amino acid:DNA transferase) (AADT) (gp46)
1 entry
DNA alpha-glucosyltransferase (AGT) (Alpha-GT) (EC 2.4.1.26)
1 entry
DNA beta-glucosyltransferase (BGT) (Beta-GT) (EC 2.4.1.27)
1 entry
5-hmdU DNA kinase 1 (5-hydroxymethyluracil DNA kinase) (P-loop kinase) (gp67)
1 entry
5-hmdU DNA kinase 2 (5-hydroxymethyluracil DNA kinase) (P-loop kinase) (gp243)
4 entries
5-hmdU DNA kinase (5-hydroxymethyluracil DNA kinase) (P-loop kinase) (gp186)
2 entries
Methylcarbamoylase mom (Adenine modification enzyme mom) (Gene product 55) (gp55) (Gene product mom) (gpMom) (Modification of Mu) (Mom)
1 entry
Orphan methyltransferase M.SPBetaI (M.SPBetaI) (EC 2.1.1.37) (Cytosine-specific methyltransferase SPBetaI) (Modification methylase SPBetaI)
1 entry
Orphan methyltransferase M.Phi3TI (M.Phi3TI) (EC 2.1.1.37) (Cytosine-specific methyltransferase Phi3TI) (Modification methylase Phi3TI)
1 entry
Orphan methyltransferase M.Rho11sI (M.Rho11sI) (EC 2.1.1.37) (Bsu P11s) (Cytosine-specific methyltransferase Rho11sI) (Modification methylase Rho11sI)
1 entry
Orphan methyltransferase M.SPRI (M.SPRI) (EC 2.1.1.37) (Cytosine-specific methyltransferase SPRI) (Modification methylase SPRI)
1 entry
Protein Ocr (Gene product 0.3) (Gp0.3) (Overcome classical restriction) (Ocr)
2 entries
Glycyl-dTMP PLP-dependent decarboxylase (gp52)
1 entry
Putrescinyltransferase (Amino acid:DNA transferase) (AADT)
3 entries
Restriction inhibitor protein ral (Antirestriction protein)
1 entry
Radical SAM Nalpha-GlyT isomerase (gp53) (rSAM glycinyl-thymine isomerase)
1 entry
Serinyltransferase (Amino acid:DNA transferase) (AADT) (gp247)
1 entry
