Ribosomal frameshifting (kw:KW-0688)
Programmed Ribosomal frameshifting  is an alternate mechanism of translation to merge proteins encoded by two overlapping open reading frames. The frameshift occurs at low frequency and consists of ribosomes slipping by one base in either the 5'(-1) or 3'(+1) directions during translation. Some viruses contains both a +1 and a -1 ribosomal frameshifts
is an alternate mechanism of translation to merge proteins encoded by two overlapping open reading frames. The frameshift occurs at low frequency and consists of ribosomes slipping by one base in either the 5'(-1) or 3'(+1) directions during translation. Some viruses contains both a +1 and a -1 ribosomal frameshifts
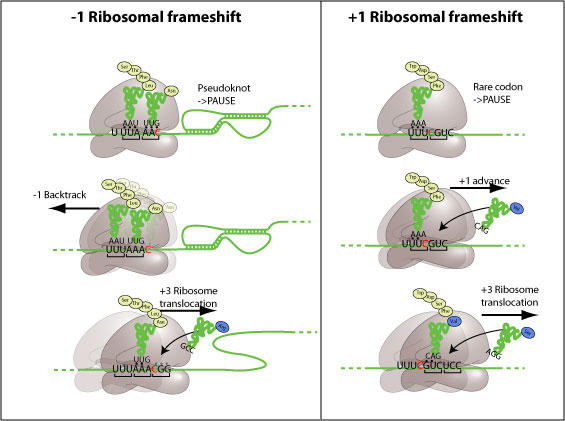
-1 Ribosomal frameshift
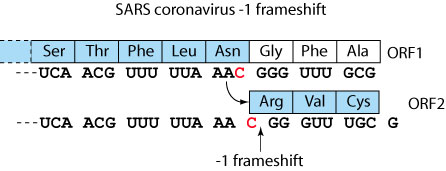
All cis-acting frameshift signals encoded in mRNAs are minimally composed of two functional elements: a heptanucleotide "slippery sequence" conforming to the general form X XXY YYZ, followed by a RNA structural element, usually a H-type RNA pseudoknot, positioned an optimal number of nucleotides (5 to 9) downstream. Mechanistically, the ribosome is stalled on the slippery sequence by the pseudoknot structure in 3'. In about 10% cases, the ribosome will backtrack one nucleotide, creating one or more mismatchs between tRNAs and mRNA. Translation resolves on the backtracked ribosome in the -1 frame.
| Family | Genus | Virus | Type | RefSeq | .......... XXXYYYZ .......... | Frameshift position | Freq | Ref |
| -1/-2 FRAMESHIFTS | ||||||||
| Totiviridae | Giardiavirus | Giardia lamblia virus | -1 | NC_003555 | CGUGCGCCAU CCCUUUA UCCGAUCGUG | 2871 |  | |
| Totivirus | Saccharomyces cerevisiae virus L-A | -1 | NC_003745 | GUACUCAGCA GGGUUUA GGAGUGGUAG | 1964 |  |
||
| Astroviridae | Avastrovirus | Turkey astrovirus 1 | -1 | NC_002470 | CUACGUGUUC AAAAAAC UAGAUAGUCA | 3307 |  |
|
| Mamastrovirus | Human astrovirus 1 | -1 | NC_001943 | ACAAGGCCCC AAAAAAC UACAAAGGGC | 2845 |  |
||
| Luteoviridae | Enamovirus | Pea enation mosaic virus-1 | -1 | NC_003629 | CCAGACGCUC GGGAAAC GGAUUAUUCC | 2035 |  |
|
| Luteovirus | Barley yellow dwarf virus-PAV | -1 | NC_004750 | TTGACTCTGT GGGUUUU AGAGGGGCTC | 1159 |  |
||
| Polerovirus | Potato leafroll virus | -1 | NC_001747 | CAAACAAGCC UUUAAAU GGGCAAGCGG | 1774 |  |
||
| Arteriviridae | Arterivirus | Equine arteritis virus | -1 | NC_002532 | CAGUGAAUCA GUUAAAC UGAGAGCGCC | 5405 |  |
|
| Porcine reproductive and respiratory syndrome virus | -2 | NC_001961 | UGAGCCGUCA GGUUUUU GACCUCGUCU | 3887-3888 |  |
|||
| Coronaviridae | Alphacoronavirus | Human coronavirus 229E | -1 | NC_002645 | AUAACAGUUA UUUAAAC GAGUCCGGGG | 12520 |  |
|
| Betacoronavirus | Human SARS coronavirus | -1 | NC_004718 | CAUCAACGUU UUUAAAC GGGUUUGCGG | 13398 |  |
||
| Gammacoronavirus | Avian Infectious bronchitis virus | -1 | NC_002645 | AUAAGAAUUA UUUAAAC GGGUACGGGG | 12354 |  |
||
| Bafinivirus | White bream virus | -1 | NC_008516 | AGCAGCCGCC UUUAAAC TGGAGGGCAG | 14555 | - | ||
| Torovirus | Breda virus | -1 | NC_007447 | CUGUUGGUGA UUUAAAC UGCUGAGAGA | 14184 | - | ||
| FLaviviridae | Flavivirus | West nile virus | -1 | NC_009942 | UAUGAUUGAC CCUUUUC AGUUGGGCCU | 3552 | 30-50% |  |
| Mesoniviridae | Mesonivirus | Alphamesonivirus 1 | -1 | NC_015668 | GCACCCUUUG GAUUUUC CAAAGUGGGC | 7841 | 
|
|
| Picornaviridae | Cardiovirus | Encephalomyocardidis virus | -1 | NC_001479 | CAAGGAAACA GGUUUUC CAGACCCAAG | 3998 |  |
|
| Tombusviridae | Dianthovirus | Red clover necrotic mosaic virus | -1 | NC_003756 | AAUCCCUUGA GGAUUUU UAGGCGGCGG | 831 |  |
|
| Retroviridae | Alpharetrovirus | Avian leukosis virus | -1 | NC_001408 | UCCGCUUGAC AAAUUUA UAGGGAGGGC | 2474 |  |
|
| Betaretrovirus | Mouse mammary tumor virus | -1 | NC_001503 | CUGAAAAUUC AAAAAAC UUGUAAAGGG | 2082 |  |
||
| Deltaretrovirus | Human T-lymphotropic virus 1 | -1 | NC_001436 | UCCCACACCC AAAAAAC UCCAUAGGGG | 1718 |  |
||
| Lentivirus | Human immunodeficiency virus 1 | -1 | NC_001802 | GACAGGCUAA UUUUUUA GGGAAGAUCU | 1637 |  |
||
| Unclassified | Sobemovirus | Cocksfoot mottle sobemovirus | -1 | NC_002618 | CAAUCCGGCC UUUAAAC UACCAGCGGG | 1640 |  |
|
| Umbravirus | Groundnut rosette virus | -1 | NC_003603 | CCGGGGCACA AAAUUUU UAGUUGGGGA | 867 |  |
||
| Myoviridae | Mulikevirus | Mu phage | -2 | NC_000929 | CGCUGACACA AGAACGG GGGCGAGUGG | 23960 |  |
|
| Myoviridae | P2likeviruses | Enterobacteria phage P2 | -1 | NC_003603 | CGGUAAGGUG GUCGGUU UUUUGUCGCC | 19679 |  |
|
| Siphoviridae | Lambdalikevirus | Phage lambda | +1 | NC_001416 | AGAGCCUGUU UCUGCGG GAAAGTGTTC | 10110 |  |
|
| Togaviridae | Alphavirus | Sindbis virus | +1 | NC_001547 | GCUGCCUGCC UUUUUUA GUGGUUCCGG | 10028 | 10-18% |  |
Source:Michael Bekaert, thesis (in french) etude du decalage de phase de lecture dans le genome de Saccharomyces cerevisiae
+1 ribosomal frameshift
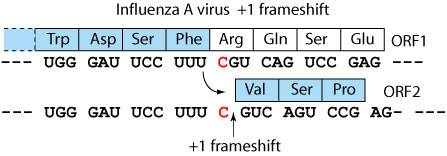
This frameshift is mechanistically different from the -1. The ribosome stalls on a slippery sequence, making a long pause at a rare codon for which few tRNAs are available. This long pause can be resolved by a movement forward of one nucleotide, creating one or more mismatchs between tRNAs and mRNA. Translation resolves on the advanced ribosome in the +1 frame.
| Family | Genus | Virus | Type | RefSeq | .......... XXXYYYZ .......... | *{color:white}Frameshift position | *{color:white}Freq | *{color:white}Ref |
| +1 FRAMESHIFTS (rare codon in yellow) | ||||||||
| Orthomyxoviridae | Influenzavirus A | Influenzavirus A | +1 | NC_002022 | UCUGGGAUUC CUUUCGU CAGUCCGAG | 599 |  |
|
| Totiviridae | Leishmaniavirus | Leishmania RNA virus 1-4 | +1 | NC_003601 | Not Identified | NA |  |
|
| Siphoviridae | Lambdalikevirus | Bacillus phage SPP1 | +1 | NC_004166 | ACTGTTCCCG CTCCCC TAATGCGCC | 11,111 | ~10% |  Origin and function of the two major tail proteins of bacteriophage SPP1 Isabelle Auzat, Anja Dr?ge, Frank Weise, Rudi Lurz, Paulo Tavares Mol. Microbiol. November 2008; 70: 557-569 |
Florian Zirkel, Andreas Kurth, Phenix-Lan Quan, Thomas Briese, Heinz Ellerbrok, Georg Pauli, Fabian H. Leendertz, W. Ian Lipkin, John Ziebuhr, Christian Drosten, Sandra Junglen
MBio 2011; 2: e00077-00011
Ezequiel Balmori Melian, Edward Hinzman, Tomoko Nagasaki, Andrew E. Firth, Norma M. Wills, Amanda S. Nouwens, Bradley J. Blitvich, Jason Leung, Anneke Funk, John F. Atkins, Roy Hall, Alexander A. Khromykh
J. Virol. February 2010; 84: 1641?1647
Andrew E. Firth, Betty Yw Chung, Marina N. Fleeton, John F. Atkins
Virol. J. 2008; 5: 108
Ezequiel Balmori Melian, Edward Hinzman, Tomoko Nagasaki, Andrew E. Firth, Norma M. Wills, Amanda S. Nouwens, Bradley J. Blitvich, Jason Leung, Anneke Funk, John F. Atkins, Roy Hall, Alexander A. Khromykh
J. Virol. February 2010; 84: 1641?1647
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)0 entry grouped by protein
