Flaviviridae (taxid:11050)
More on Zika virus here
VIRION
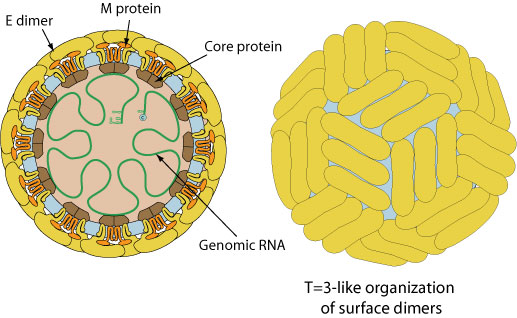
Enveloped, spherical, about 50 nm in diameter. The surface proteins are arranged in an icosahedral-like symmetry.
Source: Zhang et al

GENOME
Monopartite, linear, ssRNA(+) genome of about 9.7-12 kb. The genome 3' terminus is not polyadenylated but forms a loop structure. The 5' end has a methylated nucleotide cap (to allow translation) or a genome-linked protein (VPg).
GENE EXPRESSION
The virion RNA is infectious and serves as both the genome and the viral messenger RNA. The whole genome is translated into a polyprotein, which is processed co- and post-translationally by host and viral proteases.
ENZYMES
- RNA-dependent RNA polymerase [NS5]
- Cell-type capping
- Polyprotein major protease (Peptidase S7, S29 or S31) [NS3]
REPLICATION
CYTOPLASMIC
- Attachement of the viral envelope protein E to host receptors mediates internalization into the host cell by clathrin-mediated endocytosis or by apoptotic mimicry
- Fusion of virus membrane with host endosomal membrane. RNA genome is released into the cytoplasm.
- The positive-sense genomic ssRNA is translated into a polyprotein, which is cleaved into all structural and non structural proteins (to yield the replication proteins).
- Replication takes place at the surface of endoplasmic reticulum in cytoplasmic viral factories. A dsRNA genome is synthesized from the genomic ssRNA(+).
- The dsRNA genome is transcribed/replicated thereby providing viral mRNAs/new ssRNA(+) genomes.
- Virus assembly occurs at the endoplasmic reticulum and seems to be facilitated by the viral ionic channel. The virion buds at the endoplasmic reticulum and is transported to the Golgi apparatus.
- Release of new virions by exocytosis.
Host-virus interaction
Adaptive immune response inhibition
Dengue NS1 protein inhibits host complement by binding C4 and reducing C4b deposition and C3 convertase activity. Through this mechanism, NS1 protects DENV from complement-dependent neutralization in solution.
 .
.
Apoptosis modulation
Flaviviruses NS3 induces apoptosis through host caspases activation


 .
.
Autophagy modulation
Flaviviruses NS4A and NS4B induces autophagy signaling

 .
.
Cell-cycle modulation
Hepatitis C virus induces down-regulation of the retinoblastoma tumor suppressor and G1/S host cell cycle checkpoint dysregulation
 .
.
Innate immune response inhibition
Hepatitis C virus inhibits the interferon signaling pathway by blocking the IFN receptors  , STAT1 and TYK2
, STAT1 and TYK2
 , whilst
Dengue virus blocks STAT2
, whilst
Dengue virus blocks STAT2 
Hepatitis C virus inhibits the host IFN-mediated response by blocking host MAVS  , TRAFs
, TRAFs  and TBK1-IKBKE-DDX3 complex
and TBK1-IKBKE-DDX3 complex  , whilst
pestiviruses N(pro) blocks host IRF-3
, whilst
pestiviruses N(pro) blocks host IRF-3
 .
.
Host gene expression shutoff by virus
Hepatitis C virus activates PKR about 12h p.i. to shutoff host translation through the PKR-mediated phosphorylation of the eIF2alpha initiation factor  .
.
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)0 entry grouped by strain
Anopheles flavivirus variant 1 taxid:1903340
| Protein | ModelArchive |
| Fifo | ma-jd-viral-56717 |
