Degradation of host cell envelope components during virus entry (kw:KW-1235)
At the onset of the infective process, bacterial viruses are usually confronted with the penetration of a complex host cell envelope. Gram-negative bacterial envelope usually comprises a cell membrane, a thin periplasmic peptidoglcan cell wall covered with an outer membrane containing lipopolysaccharides (LPS). By contrast, Gram-positive bacterial envelope lacks the outer membrane, but has a much thicker peptidoglycan layer containing cell-wall glycopolymers. Moreover, these cell-envelope structures can be surrounded by other protective structures such as capsular polysaccharides (e.g. α2,8-linked polysialic acid (PSA), Hyaluronic acid (HA)), S-layer proteins or mucolic acids.
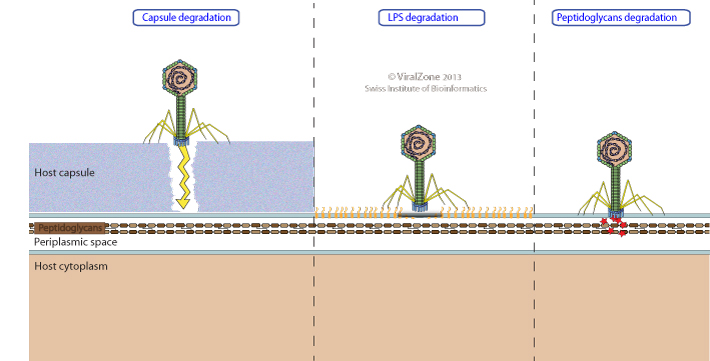
Bacterial viruses must be equipped to be able to penetrate through all these layers. Virions carry the necessary enzymatic activities to perforate the capsule, slime and peptidoglycan layers of their specific host. LPS-binding virions usually degrade these polysaccharides chains after attachment.
| Virus | Family | Host bacteria | Surface component type | Phage degrading enzyme | ref. |
| Phage φ92 | Myoviridae | E.coli (K1, K5, K92), Salmonella strains | PSA, capsular | Endosialidase gp143 |  |
| Phage K1A | Podoviridae | Escherichia coli | PSA, capsular | Endosialidase |  |
| Phage K1E | Podoviridae | Escherichia coli | PSA, capsular | Endosialidase |   |
| Phage K1F | Podoviridae | Escherichia coli | PSA, capsular | Endosialidase |   |
| Phage 63D | Siphoviridae | - | PSA, capsular | Endosialidase |   |
| Phage K1-5 | Podoviridae | Escherichia coli | PSA, capsular | Endosialidase |  |
| Phage CUS-3 | Podoviridae | Escherichia coli | PSA, capsular | Endosialidase |  |
| Phage PT-6 | unclassified | Pseudomonas aeruginosa | Alginate, capsular | Alginate lyase |  |
| Phage H4489A | unclassified | Streptococcus pyogenes | HA, capsular | Hyaluronidase |  |
| Phage 370.1 | Siphoviridae | Streptococcus pyogenes | HA, capsular | HylP1, HylP2, HylP3 |  |
| Phage H10403 | unclassified | Streptococcus pyogenes | HA, capsular | Hyaluronoglucosaminidase |  |
| Phage AF | Podoviridae | Pseudomonas putida | EPS, capsular | EPS hydrolase |  |
| Phage P27 | - | Salmonella typhimurium | LPS | Endorhamnosidase |  |
| Phage 9NA | - | Salmonella typhimurium | LPS | Endorhamnosidase |  |
| Phage KB-1 | - | Salmonella typhimurium | LPS | Endorhamnosidase |  |
| Phage Det7 | Myoviridae | Salmonella typhimurium | LPS | Endorhamnosidase |  |
| Phage Sf6 | Podoviridae | Shigella flexneri | LPS | Endorhamnosidase |  |
| Phage ε15 | Podoviridae | LPS | Endorhamnosidase |  |
|
| Phage HK620 | Podoviridae | Escherichia coli H | LPS | Tail spike endo-N-acetylglucosaminidase |  |
| Phage GA-1 | Podoviridae | Bacillus G1R | Peptidoglycan | gp3 |  |
| Phage SPβ | Siphoviridae | Bacillus subtilis | Peptidoglycan | YomI |  |
| Phage SPO1 | Myoviridae | Bacillus subtilis | Peptidoglycan | - |  |
| Phage SPP1 | Siphoviridae | Bacillus subtilis | Peptidoglycan | - |  |
| Phage M2 | Podoviridae | Bacillus subtilis | Peptidoglycan | gp3 |  |
| Phage φ29 | Podoviridae | Bacillus subtilis | Peptidoglycan | gp3 |  |
| Phage PRD1 | Tectiviridae | Escherichia coli | Peptidoglycan | p7 |  |
| Phage T3 | Podoviridae | Escherichia coli | Peptidoglycan | gp16 |  |
| Phage K1-5 | Podoviridae | Escherichia coli | Peptidoglycan | orf35 |  |
| PSA, capsular | Endosialidase |  |
|||
| Phage N4 | Podoviridae | Escherichia coli | Peptidoglycan | - |  |
| Phage T4 | Podoviridae | Escherichia coli | Peptidoglycan | gp5 |  |
| Phage T5 | Siphoviridae | Escherichia coli | Peptidoglycan | Pb2 |  |
| Phage C1 | Podoviridae | Escherichia coli | Peptidoglycan | - |  |
| Phage φ949 | - | Lactococcus lactis | Peptidoglycan | - |  |
| Phage φ31 | Siphoviridae | Lactococcus lactis | Peptidoglycan | - |  |
| Phage φc2 | Myoviridae | Lactococcus lactis | Peptidoglycan | - |  |
| Phage φr1t | - | Lactococcus lactis | Peptidoglycan | - |  |
| Phage φ6 | Cystoviridae | Pseudomonas syringae | Peptidoglycan | P5 |  |
| Phage φ13 | Cystoviridae | Pseudomonas syringae | Peptidoglycan | - |  |
| Phage SP6 | Podoviridae | Salmonella typhimurium | Peptidoglycan | orf35 |  |
| Phage P22 | Podoviridae | Salmonella typhimurium | Peptidoglycan | gp4 |  |
| LPS | Endorhamnosidase |  |
|||
| Phage φYeO3-12 | Podoviridae | Yersinia enterocolitica | Peptidoglycan | gp16 |  |
| Phage φKMV | Podoviridae | Pseudomonas aeruginosa | Peptidoglycan | gp36 |  |
| Phage φKZ | Myoviridae | Pseudomonas aeruginosa | Peptidoglycan | gp181 |   |
| Phage T7 | Podoviridae | Escherichia coli | Peptidoglycan | gp16 |  |
Michael Moak, Ian J Molineux
Mol. Microbiol. February 2004; 51: 1169-1183
Lorena Rodriguez-Rubio, Beatriz Martinez, David M Donovan, Ana Rodriguez, Pilar Garcia
Crit. Rev. Microbiol. November 2013; 39: 427-434
Yves Briers, Konstantin Miroshnikov, Oleg Chertkov, Alexei Nekrasov, Vadim Mesyanzhinov, Guido Volckaert, Rob Lavigne
Biochem. Biophys. Res. Commun. October 3, 2008; 374: 747-751
Jianlong Yan, Jiaoxiao Mao, Jianping Xie
BioDrugs December 19, 2013;
Kateryna Bazaka, Russell J Crawford, Evgeny L Nazarenko, Elena P Ivanova
Adv. Exp. Med. Biol. 2011; 715: 213-226
R Gerardy-Schahn, A Bethe, T Brennecke, M Muhlenhoff, M Eckhardt, S Ziesing, F Lottspeich, M Frosch
Mol. Microbiol. May 1995; 16: 441-450
Katharina Stummeyer, David Schwarzer, Heike Claus, Ulrich Vogel, Rita Gerardy-Schahn, Martina Muhlenhoff
Mol. Microbiol. June 2006; 60: 1123-1135
T Glonti, N Chanishvili, P W Taylor
J. Appl. Microbiol. February 2010; 108: 695-702
Martina Muhlenhoff, Katharina Stummeyer, Melanie Grove, Markus Sauerborn, Rita Gerardy-Schahn
J. Biol. Chem. April 11, 2003; 278: 12634-12644
Anneleen Cornelissen, Pieter-Jan Ceyssens, Victor N Krylov, Jean-Paul Noben, Guido Volckaert, Rob Lavigne
Virology December 20, 2012; 434: 251-256
Stefanie Barbirz, Jurgen J Muller, Charlotte Uetrecht, Alvin J Clark, Udo Heinemann, Robert Seckler
Mol. Microbiol. July 2008; 69: 303-316
R Wollin, U Eriksson, A A Lindberg
J. Virol. June 1981; 38: 1025-1033
Monika Walter, Christian Fiedler, Renate Grassl, Manfred Biebl, Reinhard Rachel, X Lois Hermo-Parrado, Antonio L Llamas-Saiz, Robert Seckler, Stefan Miller, Mark J van Raaij
J. Virol. March 2008; 82: 2265-2273
J E Chua, P A Manning, R Morona
Microbiology (Reading, Engl.) July 1999; 145 ( Pt 7): 1649-1659
K Takeda, H Uetake
Virology March 1973; 52: 148-159
David Schwarzer, Falk F. R. Buettner, Christopher Browning, Sergey Nazarov, Wolfgang Rabsch, Andrea Bethe, Astrid Oberbeck, Valorie D. Bowman, Katharina Stummeyer, Martina M?hlenhoff, Petr G. Leiman, Rita Gerardy-Schahn
J. Virol. October 2012; 86: 10384?10398
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)64 entries grouped by protein
13 entries
Depolymerase, capsule K1-specific (EC 4.-.-.-) (Dep_kpv71) (Gene product 52) (gp52) (Probable tail spike protein)
1 entry
Depolymerase, capsule K11-specific (Probable tail fiber protein)
1 entry
Depolymerase, capsule KN4-specific (Probable tail spike protein)
1 entry
Depolymerase, capsule K21-specific (Probable tail spike protein)
1 entry
Depolymerase, capsule KN5-specific (Probable tail fiber protein)
1 entry
Depolymerase, capsule K25-specific (Probable tail fiber protein)
1 entry
Probable tail spike protein (Depolymerase, capsule K35-specific)
1 entry
Depolymerase, capsule K1-specific (Probable tail fiber protein)
1 entry
Depolymerase, capsule K64-specific (Probable tail fiber protein)
1 entry
Depolymerase, capsule K30/K69-specific (Probable tail spike protein)
5 entries
Depolymerase 1, capsule K3-specific (Gene product 37) (gp37) (KP32gp37) (Probable tail spike protein)
5 entries
Depolymerase 2, capsule K5-specific (Gene product 38) (gp38) (K5 depolymerase) (Probable tail spike protein)
10 entries
Exolysin (EC 4.2.2.-)
3 entries
Pre-neck appendage protein (Gene product 12) (gp12) (Late protein GP12) (NP1) (Protein p12)
6 entries
Tail spike protein (Depolymerase, capsule K5-specific) (EC 4.-.-.-)
2 entries
Morphogenesis protein 1 (Gene product 13) (gp13) (Protein p13)
1 entry
Hyaluronoglucosaminidase (Hyaluronidase) (EC 3.2.1.35)
1 entry
Hyaluronoglucosaminidase (Hyaluronidase) (EC 3.2.1.35)
1 entry
Pre-baseplate central spike protein Gp5 (Pre-Gp5) (Peptidoglycan hydrolase gp5) (EC 3.2.1.17)
4 entries
DNA terminal protein (Gene product 3) (gp3) (Protein p3)
2 entries
Probable tape measure protein (TMP) (Transglycosylase) (EC 4.2.2.n1)
2 entries
