Degradation of host chromosome by virus (kw:KW-1247)
The rapid synthesis of viral DNA necessitates access to a large pool of nucleoside 5'-triphophates. Some prokaryotic viruses degrade the host chromosome at the onset of infection. The breakdown products (nucleosides) can be reused to synthesize viral DNA (e.g. T2, T4, T6) or can be excreted from the host cell (e.g. T5) 
 .
.
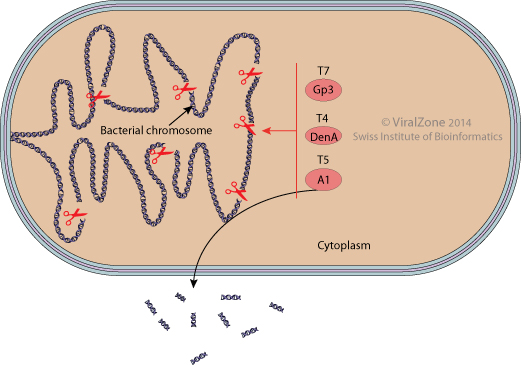
T7 for example encodes an endonuclease and a 5'->3' exonuclease required for host chromosome degradation. More than 80% of nucleotides used in synthesizing the DNA of T7 progeny derive from the host chromosome 
 .
.
Viruses that use this strategy must evolve a mechanism by which they can distinguish their own genetic material from that of their host. In the case
of bacteriophage T4, a chemical modification of the viral cytidines by a viral specific enzyme provides a simple mechanism for distinguishing between the DNA of the host and the DNA of the phage T4.
Degrading host chromosome provides an advantage during the virus life cycle by reducing or eliminating competing host macromolecular synthesis and shutting off host gene expression. However, host RNAs are not degraded by this mean and host protein synthesis can still go on.
D P Snustad, C J Bursch, K A Parson, S H Hefeneider
J. Virol. April 1976; 18: 268-288
A Souther, R Bruner, J Elliott
J. Virol. November 1972; 10: 979-984
I B Powell, D L Tulloch, A J Hillier, B E Davidson
J. Gen. Microbiol. May 1992; 138: 945-950
N Panayotatos, A Fontaine
J. Biol. Chem. March 10, 1985; 260: 3173-3177
T J Mozer, R B Thompson, S M Berget, H R Warner
J. Virol. November 1977; 24: 642-650
H R Warner, R F Drong, S M Berget
J. Virol. February 1975; 15: 273-280
M S Center, F W Studier, C C Richardson
Proc. Natl. Acad. Sci. U.S.A. January 1970; 65: 242-248
Jianbin Wang, Yan Jiang, Myriam Vincent, Yongqiao Sun, Hong Yu, Jing Wang, Qiyu Bao, Huimin Kong, Songnian Hu
Virology February 5, 2005; 332: 45-65
Matching UniProtKB/Swiss-Prot entries
(all links/actions below point to uniprot.org website)9 entries grouped by protein
1 entry
Protein A1
1 entry
DNA-(apurinic or apyrimidinic site) endonuclease (EC 3.1.-.-) (AP endonuclease) (Apurinic-apyrimidinic endonuclease) (Protein 015)
1 entry
Endonuclease II (EC 3.1.21.8)
1 entry
Endonuclease I (EC 3.1.21.2) (Gene product 3) (Gp3) (Junction-resolving enzyme gp3)
2 entries
Exonuclease subunit 1 (EC 3.1.11.-) (Gene product 47) (gp47)
2 entries
Exonuclease subunit 2 (EC 3.1.11.-) (Gene product 46) (gp46)
1 entry
